The Regulation of Ontogenetic Diversity in Papaveraceae Compound Leaf Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
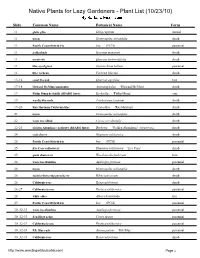
Native Plants for Lazy Gardeners - Plant List (10/23/10)
Native Plants for Lazy Gardeners - Plant List (10/23/10) Slide Common Name Botanical Name Form 11 globe gilia Gilia capitata annual 11 toyon Heteromeles arbutifolia shrub 11 Pacific Coast Hybrid iris Iris (PCH) perennial 11 goldenbush Isocoma menziesii shrub 11 scrub oak Quercus berberidifolia shrub 11 blue-eyed grass Sisyrinchium bellum perennial 11 lilac verbena Verbena lilacina shrub 13-16 coast live oak Quercus agrifolia tree 17-18 Howard McMinn man anita Arctostaphylos 'Howard McMinn' shrub 19 Philip Mun keckiella (RSABG Intro) Keckiella 'Philip Munz' ine 19 woolly bluecurls Trichostema lanatum shrub 19-20 Ray Hartman California lilac Ceanothus 'Ray Hartman' shrub 21 toyon Heteromeles arbutifolia shrub 22 western redbud Cercis occidentalis shrub 22-23 Golden Abundance barberry (RSABG Intro) Berberis 'Golden Abundance' (MAHONIA) shrub 2, coffeeberry Rhamnus californica shrub 25 Pacific Coast Hybrid iris Iris (PCH) perennial 25 Eve Case coffeeberry Rhamnus californica '. e Case' shrub 25 giant chain fern Woodwardia fimbriata fern 26 western columbine Aquilegia formosa perennial 26 toyon Heteromeles arbutifolia shrub 26 fuchsia-flowering gooseberry Ribes speciosum shrub 26 California rose Rosa californica shrub 26-27 California fescue Festuca californica perennial 28 white alder Alnus rhombifolia tree 29 Pacific Coast Hybrid iris Iris (PCH) perennial 30 032-33 western columbine Aquilegia formosa perennial 30 032-33 San Diego sedge Carex spissa perennial 30 032-33 California fescue Festuca californica perennial 30 032-33 Elk Blue rush Juncus patens '.l1 2lue' perennial 30 032-33 California rose Rosa californica shrub http://www weedingwildsuburbia com/ Page 1 30 032-3, toyon Heteromeles arbutifolia shrub 30 032-3, fuchsia-flowering gooseberry Ribes speciosum shrub 30 032-3, Claremont pink-flowering currant (RSA Intro) Ribes sanguineum ar. -

© 2020 Theodore Payne Foundation for Wild Flowers & Native Plants. No
April 24, 2020 Theodore Payne Foundation’s Wild Flower Hotline is made possible by donations, memberships and sponsors. You can support TPF by shopping the online gift store as well. A new, pay by phone, contactless plant pickup system is now available. Details here. Widespread closures remain in place. If you find an accessible trail, please practice social distancing precautions. The purpose for the Wild Flower Hotline now is NOT to send you out to localities to view wild flowers, but to post photos that assure you—virtually—that California’s wild spaces are still open for business for flowers and their pollinators. This week Mother Nature turned on the furnace and with the hot temperatures, our spring love fest with the beloved California poppy will soon come to an end. Throughout the state, poppies are setting both seed and promise for a glorious Spring 2021. Antelope Valley and the surrounding area has a great display of luminous orange California poppies (Eschscholzia californica), electric yellow monolopia (Monolopia lanceolata) and patches of goldfields (Lasthenia sp.). Spotted among the overwhelming yellow-orange color, are lupine (Lupinus spp.), tansy leafed phacelia (Phacelia tanacetifolia) and popcorn flower (Cryptantha spp.). You do not need to leave your home to see the poppies at the Antelope Valley State Poppy Reserve. Just view the live stream online at the preserve via the PoppyCam. Poppies (Eschscholzia californica) in Antelope Valley. Photo by Don Vogt © 2020 Theodore Payne Foundation for Wild Flowers & Native Plants. No reproduction of any kind without written permission. Native plants are blooming and abundant in a South Pasadena nature park. -

Draft Plant Propagation Protocol
Plant Propagation Protocol for Cardamine californica ESRM 412 – Native Plant Production Protocol URL: https://courses.washington.edu/esrm412/protocols/CACA39.pdf TAXONOMY Plant Family Scientific Name Brassicaceae/Cruciferae Common Name Mustards Species Scientific Name Scientific Name Cardamine californica (Nutt.) Greene Varieties Cardamine californica (Nutt.) Greene var. californica Cardamine californica (Nutt.) Greene var. cardiophylla (Greene) Rollins Cardamine californica (Nutt.) Greene var. cuneate (Greene) Rollins Cardamine californica (Nutt.) Greene var. integrifolia (Nutt.) Rollins Cardamine californica (Nutt.) Greene var. sinuata (Greene) O.E. Schulz Sub-species N/A Cultivar N/A Common Synonym(s) Dentaria californica Nutt. Dentaria californica Nutt. var. cardiophylla (Greene) Detling Dentaria californica Nutt. var. cuneata (Greene) Detling Dentaria californica Nutt. var. integrifolia (Nutt.) Detling Dentaria californica Nutt. var. sinuata (Greene) Detling Common Name(s) Milkmaids, bitter cress, toothwort Species Code (as per USDA Plants CACA39 database) GENERAL INFORMATION Geographical range Distribution in North America includes California, Oregon, and Washington6 Ecological distribution This plant occurs in many different communities including Foothill Woodland, Redwood Forest, Mixed Evergreen Forest, Coastal Prairie, Northern Oak Woodland.3 They grow in woodland or shaded areas and moist woods. Climate and elevation range They grow from 0 to 2770 meters in elevation.3 Local habitat and abundance Milkmaids grow in open meadows, -

Phylogeography of a Tertiary Relict Plant, Meconopsis Cambrica (Papaveraceae), Implies the Existence of Northern Refugia for a Temperate Herb
Article (refereed) - postprint Valtueña, Francisco J.; Preston, Chris D.; Kadereit, Joachim W. 2012 Phylogeography of a Tertiary relict plant, Meconopsis cambrica (Papaveraceae), implies the existence of northern refugia for a temperate herb. Molecular Ecology, 21 (6). 1423-1437. 10.1111/j.1365- 294X.2012.05473.x Copyright © 2012 Blackwell Publishing Ltd. This version available http://nora.nerc.ac.uk/17105/ NERC has developed NORA to enable users to access research outputs wholly or partially funded by NERC. Copyright and other rights for material on this site are retained by the rights owners. Users should read the terms and conditions of use of this material at http://nora.nerc.ac.uk/policies.html#access This document is the author’s final manuscript version of the journal article, incorporating any revisions agreed during the peer review process. Some differences between this and the publisher’s version remain. You are advised to consult the publisher’s version if you wish to cite from this article. The definitive version is available at http://onlinelibrary.wiley.com Contact CEH NORA team at [email protected] The NERC and CEH trademarks and logos (‘the Trademarks’) are registered trademarks of NERC in the UK and other countries, and may not be used without the prior written consent of the Trademark owner. 1 Phylogeography of a Tertiary relict plant, Meconopsis cambrica 2 (Papaveraceae), implies the existence of northern refugia for a 3 temperate herb 4 Francisco J. Valtueña*†, Chris D. Preston‡ and Joachim W. Kadereit† 5 *Área de Botánica, Facultad deCiencias, Universidad de Extremadura, Avda. de Elvas, s.n. -

Vascular Plants at Fort Ross State Historic Park
19005 Coast Highway One, Jenner, CA 95450 ■ 707.847.3437 ■ [email protected] ■ www.fortross.org Title: Vascular Plants at Fort Ross State Historic Park Author(s): Dorothy Scherer Published by: California Native Plant Society i Source: Fort Ross Conservancy Library URL: www.fortross.org Fort Ross Conservancy (FRC) asks that you acknowledge FRC as the source of the content; if you use material from FRC online, we request that you link directly to the URL provided. If you use the content offline, we ask that you credit the source as follows: “Courtesy of Fort Ross Conservancy, www.fortross.org.” Fort Ross Conservancy, a 501(c)(3) and California State Park cooperating association, connects people to the history and beauty of Fort Ross and Salt Point State Parks. © Fort Ross Conservancy, 19005 Coast Highway One, Jenner, CA 95450, 707-847-3437 .~ ) VASCULAR PLANTS of FORT ROSS STATE HISTORIC PARK SONOMA COUNTY A PLANT COMMUNITIES PROJECT DOROTHY KING YOUNG CHAPTER CALIFORNIA NATIVE PLANT SOCIETY DOROTHY SCHERER, CHAIRPERSON DECEMBER 30, 1999 ) Vascular Plants of Fort Ross State Historic Park August 18, 2000 Family Botanical Name Common Name Plant Habitat Listed/ Community Comments Ferns & Fern Allies: Azollaceae/Mosquito Fern Azo/la filiculoides Mosquito Fern wp Blechnaceae/Deer Fern Blechnum spicant Deer Fern RV mp,sp Woodwardia fimbriata Giant Chain Fern RV wp Oennstaedtiaceae/Bracken Fern Pleridium aquilinum var. pubescens Bracken, Brake CG,CC,CF mh T Oryopteridaceae/Wood Fern Athyrium filix-femina var. cyclosorum Western lady Fern RV sp,wp Dryopteris arguta Coastal Wood Fern OS op,st Dryopteris expansa Spreading Wood Fern RV sp,wp Polystichum munitum Western Sword Fern CF mh,mp Equisetaceae/Horsetail Equisetum arvense Common Horsetail RV ds,mp Equisetum hyemale ssp.affine Common Scouring Rush RV mp,sg Equisetum laevigatum Smooth Scouring Rush mp,sg Equisetum telmateia ssp. -

Argemone Mexicana L.)
Electronic Journal of Biotechnology ISSN: 0717-3458 Vol.11 No.1, Issue of January 15, 2008 © 2008 by Pontificia Universidad Católica de Valparaíso -- Chile Received March 21, 2007 / Accepted Mayo 20, 2007 DOI: 10.2225/vol11-issue1-fulltext-3 SHORT COMMUNICATION Agrobacterium-mediated transient transformation of Mexican prickly poppy (Argemone mexicana L.) Gregorio Godoy-Hernández* Unidad de Bioquímica y Biología Molecular de Plantas Centro de Investigación Científica de Yucatán A.C. C. 43 No. 130, Chuburná de Hidalgo 97200 Mérida, Yucatán, México Tel: 999 9428330 Fax: 999 9 81 39 00 E-mail: [email protected] Elidé Avilés-Berzunza Unidad de Bioquímica y Biología Molecular de Plantas Centro de Investigación Científica de Yucatán A.C. C. 43 No. 130, Chuburná de Hidalgo 97200 Mérida, Yucatán, México Tel: 999 9428330 Fax: 999 9 81 39 00 E-mail: [email protected] Mildred Carrillo-Pech Unidad de Bioquímica y Biología Molecular de Plantas Centro de Investigación Científica de Yucatán A.C. C. 43 No. 130, Chuburná de Hidalgo 97200 Mérida, Yucatán, México Tel: 999 9428330 Fax: 999 9 81 39 00 E-mail: [email protected] Felipe Vázquez-Flota Unidad de Bioquímica y Biología Molecular de Plantas Centro de Investigación Científica de Yucatán A.C. C. 43 No. 130, Chuburná de Hidalgo 97200 Mérida, Yucatán, México Tel: 999 9428330 Fax: 999 9 81 39 00 E-mail: [email protected] Website: http://www.cicy.mx Financial support: Consejo Nacional de Ciencia y Tecnología (CONACYT) from México (Grant No. 28643-B). Keywords: Argemone mexicana, β-glucuronidase, benzylisoquinoline alkaloids, genetic transformation, neomycin phosphotransferase II. -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

Vascular Plant and Vertebrate Inventory of Chiricahua National Monument
In Cooperation with the University of Arizona, School of Natural Resources Vascular Plant and Vertebrate Inventory of Chiricahua National Monument Open-File Report 2008-1023 U.S. Department of the Interior U.S. Geological Survey National Park Service This page left intentionally blank. In cooperation with the University of Arizona, School of Natural Resources Vascular Plant and Vertebrate Inventory of Chiricahua National Monument By Brian F. Powell, Cecilia A. Schmidt, William L. Halvorson, and Pamela Anning Open-File Report 2008-1023 U.S. Geological Survey Southwest Biological Science Center Sonoran Desert Research Station University of Arizona U.S. Department of the Interior School of Natural Resources U.S. Geological Survey 125 Biological Sciences East National Park Service Tucson, Arizona 85721 U.S. Department of the Interior DIRK KEMPTHORNE, Secretary U.S. Geological Survey Mark Myers, Director U.S. Geological Survey, Reston, Virginia: 2008 For product and ordering information: World Wide Web: http://www.usgs.gov/pubprod Telephone: 1-888-ASK-USGS For more information on the USGS-the Federal source for science about the Earth, its natural and living resources, natural hazards, and the environment: World Wide Web:http://www.usgs.gov Telephone: 1-888-ASK-USGS Suggested Citation Powell, B.F., Schmidt, C.A., Halvorson, W.L., and Anning, Pamela, 2008, Vascular plant and vertebrate inventory of Chiricahua National Monument: U.S. Geological Survey Open-File Report 2008-1023, 104 p. [http://pubs.usgs.gov/of/2008/1023/]. Cover photo: Chiricahua National Monument. Photograph by National Park Service. Note: This report supersedes Schmidt et al. (2005). Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. -

Changing Pattern of Epidemic Dropsy in North India
Epidemic Dropsy in North India N. Sharma et al. ORIGINAL ARTICLE Changing Pattern of Epidemic Dropsy in North India NAVNEET SHARMA1,*, NAINA MOHAN 2, ASHISH BHALLA1, AMAN SHARMA1, SURJIT SINGH 1 1 The Department of Internal Medicine, Postgraduate Institute of Medical Education and Research, Chandigarh, India 2 King's College London, Strand, London, United Kingdom Abstract Background: Epidemic dropsy occurs due to ingestion of mustard oil contaminated with oil from Argemone mexicana, leading to edema and tenderness of the abdomen, upper and lower limbs. In this study, clinical profiles of patients presented with epidemic dropsy in north India are described. Methods: This was a prospective study of patients presented with epidemic dropsy to the emergency department of Nehru Hospital, during the period from March 2004 to December 2011. Inclusion criteria were patients presenting with tender bilateral pitting leg edema and dermal telangiectasia. Clinical and laboratory data of patients were entered into case record forms at the time of presentation until discharge from the hospital. Results: Leg edema was the principal symptom in our series, and was in concurrence with current literature. Erythema has only been reported in 35-82% of published series, though it was present in all of our patients. Similarly, features such as diarrhea, hepatomegaly and anemia were more frequent in our cases compared to the literature. Furthermore, pancytopenia which was documented on peripheral blood counts in 54% of our cases has never been reported before. Conclusion: Epidemic dropsy should be considered in patients presenting with progressive erythema, edema, and tenderness of the limbs who had a history of consumption of mustard oil and confirmation of Argemone oil contamination according to laboratory tests. -

Epidemic Dropsy in India
Postgrad Med J 1999;75:657–661 © The Fellowship of Postgraduate Medicine, 1999 Postgrad Med J: first published as 10.1136/pgmj.75.889.657 on 1 November 1999. Downloaded from Epidemic dropsy in India B D Sharma, Sanjay Malhotra, Vikram Bhatia, Mandeep Rathee Summary Epidemic dropsy results from ingestion of edible oil adulterated with Argemone Epidemic dropsy is a clinical state mexicana (Mexican Poppy) oil. The outbreak of epidemic dropsy in the Indian resulting from use of edible oils capital, New Delhi, during the rainy season of 1998 was of one of the most severe adulterated with Argemone mexi- forms and had repercussions in both health and political circles. Some 2552 cana oil. Sanguinarine and dehyd- cases were reported and 65 deaths occurred between 5 August and 12 October, rosanguinarine are two major causing untold misery and economic loss to the aVected families. The actual fig- toxic alkaloids of Argemone oil, ures are likely to be much higher due to nonreporting of milder cases to the hos- which cause widespread capillary pitals. The aim of this article is to consolidate and update the available dilatation, proliferation and in- information on clinical aspects of epidemic dropsy. creased capillary permeability. The condition was first reported by Lyon in 1877 from Calcutta1 and has since Leakage of the protein-rich occurred in other countries including the Fiji Islands, Mauritius, Madagascar, plasma component into the extra- South Africa and Burma (Myanmar).2 In India, it has been reported from time cellular compartment leads to the to time from the States of West Bengal, Bihar, Orissa, Madhya Pradesh, Uttar formation of oedema. -

A Systematic Review on Main Chemical Constituents of Papaver Bracteatum
Journal of Medicinal Plants A Systematic Review on Main Chemical Constituents of Papaver bracteatum Soleymankhani M (Ph.D. student), Khalighi-Sigaroodi F (Ph.D.)*, Hajiaghaee R (Ph.D.), Naghdi Badi H (Ph.D.), Mehrafarin A (Ph.D.), Ghorbani Nohooji M (Ph.D.) Medicinal Plants Research Center, Institute of Medicinal Plants, ACECR, Karaj, Iran * Corresponding author: Medicinal Plants Research Center, Institute of Medicinal Plants, ACECR, P.O.Box: 33651/66571, Karaj, Iran Tel: +98 - 26 - 34764010-9, Fax: +98 - 26-34764021 E-mail: [email protected] Received: 17 April 2013 Accepted: 12 Oct. 2014 Abstract Papaver bracteatum Lindly (Papaveraceae) is an endemic species of Iran which has economic importance in drug industries. The main alkaloid of the plant is thebaine which is used as a precursor of the semi-synthetic and synthetic compounds including codeine and naloxone, respectively. This systematic review focuses on main component of Papaver bracteatum and methods used to determine thebaine. All studies which assessed the potential effect of the whole plant or its extract on clinical or preclinical studies were reviewed. In addition, methods for determination of the main components, especially thebaine, which have been published from 1948 to March 2013, were included. Exclusion criteria were agricultural studies that did not assess. This study has listed alkaloids identified in P. bracteatum which reported since 1948 to 2013. Also, the biological activities of main compounds of Papaver bracteatum including thebaine, isothebaine, (-)-nuciferine have been reviewed. As thebaine has many medicinal and industrial values, determination methods of thebaine in P. bracteatum were summarized. The methods have being used for determination of thebaine include chromatographic (HPLC, GC and TLC) and non chromatographic methods. -

Argemone Mexicana
Argemone mexicana General description Scientific Name with Author Argemone mexicana L. Synonyms Argemone leiocarpa Greene; Argemone ochroleuca Sweet; Echtrus trivialis Lour.; Echtrus mexicanus (L.) Nieuwl.; Argemone vulgaris Spach; Argemone versicolor Salisb.; Argemone spinosa Moench; Argemone sexvalis Stokes; Argemone mucronata Dum. Cours. ex Steud.; Argemone mexicana var. typica Prain; Argemone mexicana var. parviflora Kuntze; Argemone mexicana var. ochroleuca (Sweet) Lindl.; Argemone mexicana var. lutea Kuntze; Argemone mexicana fo. leiocarpa (Greene) G.B. Ownbey (Pires, 2009). Family Papaveraceae Vernacular Names Mexican poppy, prickly poppy, yellow thistle, Mexican thistle (En). Argémone, pavot épineux, pavot du Mexique, tache de l’œil, chardon du pays (Fr) (Bosch, 2008) Botanical Description Argemone mexicana is an annual herb, growing up to 150 cm with a slightly branched tap root. Its stem is branched and usually extremely prickly. It exudes a yellow juice when cut. It has showy yellow flowers. Leaves are thistle-like and alternate, without leaf stalks (petioles), toothed (serrate) and the margins are spiny. The grey-white veins stand out against the bluish-green upper leaf surface. The stem is oblong in cross-section. Flowers are at the tips of the branches (are terminal) and solitary, yellow and of 2.5-5 cm diameter. Fruit is a prickly oblong or egg-shaped (ovoid) capsule. Seeds are very numerous, nearly spherical, covered in a fine network of veins, brownish black and about 1 m m in diameter (Nacoulma, 1996; Bosch, 2008). 1 MEAMP – Appear Project – 75 September 2012 – August 2014 Photo LABIOCA 1. Argemone mexicana Origin and Distribution Argemone mexicana is native in Mexico and the West Indies, but has become pantropical after accidental introduction or introduction as an ornamental.