The Systematic Value of Nuclear DNA Content for All Species of Narcissus L. (Amaryllidaceae)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Extended Glossary 11 Extended Glossary
Extended Glossary 11 EXTENDED GLOSSARY This glossary combines some of the helpful found on the RHS website at www.rhs.org.uk/ introductory sections from older editions in an agmplants. alphabetical listing. A fuller, more discursive account The AGM plant list has, to date, been re-examined of plant names, Guide to Plant Names, and a detailed every 10 years. The latest review was carried out guide to the typography of plant names, during 2012 and published in February 2013. Recommended Style for Printing Plant Names, are both available as RHS Advisory Leaflets. To request Botanical Names a copy of either please send an A4 sae to The Compiler at the contact address given on page 5. The aim of the botanical naming system is to provide each different plant with a single, unique, universal Advisory Committee on name. The basic unit of plant classification is the Nomenclature and Taxonomy species. Species that share a number of significant characteristics are grouped together to form a genus See under the new name of Nomenclature and (plural genera). The name of a species is made up of Taxonomy Advisory Group two elements; the name of the genus followed by the specific epithet, for example, Narcissus romieuxii. Authorities Variation within a species can be recognised by division into subspecies (usually abbreviated to In order that plant names can be used with precision subsp.), varietas (or variety abbreviated to var.) and throughout the scientific world, the name of the forma (or form abbreviated to f.). Whilst it is person who coined the name of a plant species (its unusual for a plant to have all of these, it is possible, author, or authority) is added to the plant name. -

Conserving Europe's Threatened Plants
Conserving Europe’s threatened plants Progress towards Target 8 of the Global Strategy for Plant Conservation Conserving Europe’s threatened plants Progress towards Target 8 of the Global Strategy for Plant Conservation By Suzanne Sharrock and Meirion Jones May 2009 Recommended citation: Sharrock, S. and Jones, M., 2009. Conserving Europe’s threatened plants: Progress towards Target 8 of the Global Strategy for Plant Conservation Botanic Gardens Conservation International, Richmond, UK ISBN 978-1-905164-30-1 Published by Botanic Gardens Conservation International Descanso House, 199 Kew Road, Richmond, Surrey, TW9 3BW, UK Design: John Morgan, [email protected] Acknowledgements The work of establishing a consolidated list of threatened Photo credits European plants was first initiated by Hugh Synge who developed the original database on which this report is based. All images are credited to BGCI with the exceptions of: We are most grateful to Hugh for providing this database to page 5, Nikos Krigas; page 8. Christophe Libert; page 10, BGCI and advising on further development of the list. The Pawel Kos; page 12 (upper), Nikos Krigas; page 14: James exacting task of inputting data from national Red Lists was Hitchmough; page 16 (lower), Jože Bavcon; page 17 (upper), carried out by Chris Cockel and without his dedicated work, the Nkos Krigas; page 20 (upper), Anca Sarbu; page 21, Nikos list would not have been completed. Thank you for your efforts Krigas; page 22 (upper) Simon Williams; page 22 (lower), RBG Chris. We are grateful to all the members of the European Kew; page 23 (upper), Jo Packet; page 23 (lower), Sandrine Botanic Gardens Consortium and other colleagues from Europe Godefroid; page 24 (upper) Jože Bavcon; page 24 (lower), Frank who provided essential advice, guidance and supplementary Scumacher; page 25 (upper) Michael Burkart; page 25, (lower) information on the species included in the database. -

Transversal Pattern of the Leaves of Eighteen Species of the Genus Narcissus L
Flora Montiberica 79: 28-31 (III-2021) ISSN1138-5952 – eISSN1988-799X TRANSVERSAL PATTERN OF THE LEAVES OF EIGHTEEN SPECIES OF THE GENUS NARCISSUS L. (ASPARAGALES: AMARYLLIDACEAE) IN SPAIN Pedro GÓMEZ-MURILLO1 & Irene ARELLANO-MARTÍN2 1Independent Researcher. C/Caridad, 8. planta 2, pta. 8. 29680-Estepona (Málaga, Spain) [email protected]; [email protected] ABSTRACT: Data on the pattern of the cross section of the leaves and additional information on 18 wild species of the genus Narcissus L. observed and analyzed in-situ in Spain are shown. Keywords: Daffodils; Narcissus; classification; speciation; Extremadura; Andalucía; Spain. RESUMEN: Patrón transversal de las hojas de dieciocho especies del género Narcissus L. (Asparagales: Amaryllidaceae) en España. Se muestran datos sobre patrón del corte transversal de las hojas e información adicional de 18 especies silvestres del género Narcissus L. observadas y analizadas in-situ en España. Palabras clave: Narcissus; narcisos; especiación; Clasificación; Extremadura; Andalucía; España. INTRODUCTION It is not currently listed by IUCN. The genus Narcissus L. expresses its greatest diversi- Narcissus blancoi Barra & G. López ty in the Iberian Peninsula (BLANCHARD, 1990; MAR- JAÉN: Vilches, 30SVH52, 15-02-2019. QUES & al., 2017). Currently, the real number of species Endemic species of Spain, member of Sect. Bulboco- is far from being known, due to the great variability of dii (BARRA & al., 2011). some groups. The cross section of the leaves is semicircular (fig. 1c). The taxonomy of the genus Narcissus L. has proven It is not currently listed by IUCN. to be very complex and difficult to solve (WEBB, 1980; MATHEW, 2002). Historically, the number of accepted Narcissus cantabricus DC. -

Estrategia Nacional De Conservación Y Utilización De Parientes Silvestres De Los Cultivos (PSC) Y Plantas Silvestres De Uso Alimentario (PSUA) (Borrador)
Estrategia Nacional de Conservación y Utilización de Parientes Silvestres de los Cultivos (PSC) y Plantas Silvestres de Uso Alimentario (PSUA) (Borrador) AUTORES Índice Acrónimos, abreviaturas y siglas ...........................................................................................i 1. Antecedentes ............................................................................................................... 1 2. Marco jurídico .............................................................................................................. 7 Internacional ....................................................................................................................... 7 Europeo .............................................................................................................................. 9 Nacional ............................................................................................................................ 11 3. Ámbito de aplicación .................................................................................................. 16 4. Diagnóstico de la situación actual ............................................................................... 19 Análisis de diversidad ........................................................................................................ 19 Evaluación del estado de conservación ............................................................................... 20 In situ ............................................................................................................................... -

Folia Botanica Extremadurensis, 14
Volumen 14 NOVIEMBRE 2020 FOLIA BOTANICA EXTREMADURENSIS BOTANICA FOLIA Coordinación: Francisco Mª Vázquez Pardo Secretaría: Francisco Márquez García Equipo de edición: David García Alonso, Francisco Márquez García y María José Guerra Barrena. Equipo de redacción y revisión de textos: José Blanco Salas David García Alonso Francisco Márquez García José Luis Pérez Chiscano Carlos Pinto Gomes Francisco Mª Vázquez Pardo Ilustración de portada: Ejemplar de Dactyloctenium aegyptium (L.) Willd. Edita: Centro de Investigaciones Científicas y Tecnológicas de Extremadura (CICYTEX). ISSN: 1887-6587 Depósito legal: BA-178-07 Diseño: Grupo HABITAT. Imprime: IBERPRINT. Montijo (Badajoz, España). Unidad de Biodiversidad Vegetal. Herbario HSS. Instituto de Investigaciónes Agrarias “La Orden”. A-V, km 372. 06187 GUADAJIRA (BADAJOZ (España)). Centro de Investigaciones Científicas y Tecnológicas de Extremadura (CICYTEX). Consejería de Economía, Ciencia y Agenda Digital. Junta de Extremadura. FOLIA BOTANICA EXTREMADURENSIS Vol. 14 NOVIEMBRE 2020 Coordinación: Francisco Mª Vázquez Pardo Secretaría: Francisco Márquez García Equipo de edición: David García Alonso, Francisco Márquez García, y María José Guerra Barrena. Equipo de redacción y revisi ón de textos: José Blanco Salas David García Alonso Francisco Márquez García José Luis Pérez Chiscano Carlos Pinto Gomes Francisco Mª Vázquez Pardo Ilustración de portada: Ejemplar de Dactyloctenium aegyptium (L.) Willd. Edita: Centro de Investigaciones Científicas y Tecnológicas de Extremadura (CICYTEX). ISSN: 1887-6587 Depósito legal: BA-178-07 Diseño: Grupo HABITAT. Imprime: IBERPRINT. Montijo (Badajoz, España). Unidad de Biodiversidad Vegetal. Herbario HSS. Instituto de Investigaciones Agrarias “La Orden”. A-V, km 372. 06187 GUADAJIRA (BADAJOZ (España)). Centro de Investigaciones Científicas y Tecnológicas de Extremadura (CICYTEX). Consejería de Economía, Ciencia y Agenda Digital. -

A Case Study for FSC (Forest Stewardship Council) Forests in Southwestern Spain
Article Assessment and Monitoring Protocols to Guarantee the Maintenance of Biodiversity in Certified Forests: A Case Study for FSC (Forest Stewardship Council) Forests in Southwestern Spain Antonio J. Sánchez-Almendro 1, Pablo J. Hidalgo 1,*, Rosario Galán 2, José M. Carrasco 3 and Javier López-Tirado 1 1 Departamento de Ciencias Integradas, Universidad de Huelva, 21071-Huelva, Spain; [email protected] (A.J.S.-A.); [email protected] (J.L.-T.) 2 FSC International Center GmbH, Adenauerallee 134, 53113 Bonn, Germany; [email protected] 3 Ence, Energía y Celulosa, Crtra. A-5000, km 7.5, 21007-Huelva, Spain; [email protected] * Correspondence: [email protected]; Tel.: +34-959-21-9886 Received: 4 August 2018; Accepted: 10 November 2018; Published: 14 November 2018 Abstract: (1) Biodiversity, sustainable development and nature conservation are fundamental issues nowadays. All companies, administrations, governments and international organisations take these issues into consideration. Sustainable forest management always requires a compromise between profitability and conservation and in this fragile equilibrium, forest certification plays a key scheme. This sustainable management is of great importance in the European Union (EU), with the Forest Stewardship Council playing a fundamental role in forest certification. This certification forms the basis of the ecosystem conservation and improvement strategy in Ence, Energía y Celulosa, the leading company dedicated to the production of eucalyptus in Spain; (2) A three-phase protocol (identification of High Conservation Values, assessment of conservation areas and monitoring program), has been developed, providing clear, objective criteria, particularly concerning FSC (Forest Stewardship Council) Principle 9, the primary goal being the development and application of these objective criteria in the Ence conservation areas in the province of Huelva (Spain). -

Plants Given RHS Exhibition Awards from 2008–2010
HanbUryana 6: 83–139 (2012) 83 Plants given RHS Exhibition Awards 2008–2010 C.M. WhiTehOUse & J.J. CUbey RHS Garden Wisley, Surrey GU23 6QB The following list of 416 awards has been compiled from the plant award descriptions written by Plant Committee Secretaries, or in the case of the Orchid Committee by Johan and Clare Hermans, for the period from September 2008 until the end of December 2010. Awards made prior to this that were confirmed during this period, following, for example, application or registration of a cultivar name, are also included. References to further information and catalogued herbarium specimens and images relating to plant awards are included. Anyone wishing to visit the Herbarium at Wisley, to view the herbarium specimens or images, should contact the Keeper of the Herbarium. Paintings are commissioned for many orchids that are given awards and these can be consulted by contacting the RHS Lindley Library. References such as 155D refer to colours in the RHS Colour Chart (Fifth edition, 2007).1 For enquiries regarding these awards please email plantcommittees@rhs. org.uk or telephone 0845 260 9000. These will then be forwarded to the appropriate Plant Committee Secretary or member of staff. Key AM Award of Merit BC Botanical Certificate (awarded to the species) E Exhibited by D Description DCP Description & colour photograph FCC First Class Certificate PC Certificate of Preliminary Commendation R Raised by S Submitted by § As a flowering plant for exhibition * As a hardy flowering plant for exhibition † As a tender flowering plant for exhibition ‡ As a flowering plant for cultivation in containers 1 Available from RHS Enterprises mail order ([email protected] or telephone 01483 211320). -

(Asparagales: Amaryllidaceae): Especies Endémicas De España
20210624 Notas sobre el género Narcissus Linnaeus, 1753 (Asparagales: Amaryllidaceae): Especies endémicas de España PEDRO GÓMEZ-MURILLO Independent Researcher. 29680-Estepona, Málaga, Spain. [email protected] Resumen: GÓMEZ-MURILLO (2021). Notas sobre el género Narcissus Linnaeus, 1753 (Asparagales: Amaryllidaceae): Especies endémicas de España. España es uno de los países más ricos en narcisos silvestres, el objetivo de la presente investigación es establecer el inventario de narcisos silvestres endémicos de España. Se realizaron salidas de campo y una revisión de literatura especializada. Se registraron 23 especies endémicas, de las cuales solo 5 se encuentran evaluadas por la IUCN. Palabras clave: Conservación, España, Distribución, Clasificación, Diversidad. Abstract: GÓMEZ-MURILLO (2021). Notes on the genus Narcissus Linnaeus, 1753 (Asparagales: Amaryllidaceae): Endemic species of Spain. Spain is one of the richest countries in wild daffodils, the objective of this research is to establish the inventory of wild daffodils endemic to Spain. Field trips and a review of specialized literature were carried out. 23 endemic species were registered, of which only 5 are evaluated by the IUCN. Keywords: Conservation, Spain, Distribution, Classification, Diversity. OBSERVACIONES En España podemos encontrar alrededor de 60 especies del género Narcissus Linnaeus, 1753, siendo el país con más especies en estado silvestre del mundo (Aedo, 2013; Marques et al., 2017). En los últimos años se han descrito especies nuevas en el país (Escobar, -

The Interplay Between Inflorescence Development and Function As the Crucible of Architectural Diversity
Annals of Botany Page 1 of 17 doi:10.1093/aob/mcs252, available online at www.aob.oxfordjournals.org REVIEW: PART OF A SPECIAL ISSUE ON INFLORESCENCES The interplay between inflorescence development and function as the crucible of architectural diversity Lawrence D. Harder1,* and Przemyslaw Prusinkiewicz2 1Department of Biological Sciences, University of Calgary, Calgary, Alberta, Canada T2N 1N4 and 2Department of Computer Science, University of Calgary, Calgary, Alberta, Canada T2N 1N4 * For correspondence. Email [email protected] Received: 27 July 2012 Returned for revision: 13 September 2012 Accepted: 17 October 2012 † Background Most angiosperms present flowers in inflorescences, which play roles in reproduction, primarily Downloaded from related to pollination, beyond those served by individual flowers alone. An inflorescence’s overall reproductive contribution depends primarily on the three-dimensional arrangement of the floral canopy and its dynamics during its flowering period. These features depend in turn on characteristics of the underlying branching structure (scaffold) that supports and supplies water and nutrients to the floral canopy. This scaffold is produced by devel- opmental algorithms that are genetically specified and hormonally mediated. Thus, the extensive inflorescence diversity evident among angiosperms evolves through changes in the developmental programmes that specify http://aob.oxfordjournals.org/ scaffold characteristics, which in turn modify canopy features that promote reproductive performance in a par- ticular pollination and mating environment. Nevertheless, developmental and ecological aspects of inflorescences have typically been studied independently, limiting comprehensive understanding of the relations between inflor- escence form, reproductive function, and evolution. † Scope This review fosters an integrated perspective on inflorescences by summarizing aspects of their devel- opment and pollination function that enable and guide inflorescence evolution and diversification. -

Revisión De Los Géneros Thymbra L
Volumen 7 Julio 2013 FOLIA BOTANICA EXTREMADURENSIS Coordinación: Francisco Mª Vázquez Pardo Secretaría: Francisco Márquez García Equipo de edición: David García, María José Guerra, José Blanco y Francisco Márquez Equipo de redacción y revisión de textos: José Blanco Salas David García Alonso Francisco Márquez García José Luis Pérez Chiscano Carlos Pinto Gomes Francisco M. Vázquez Pardo Ilustración de portada: Narcissus bulbocodium L. subsp. bulbocodium var. obesus (Salib.) Baker Edita: Consejería de Empleo, Empresa e Innovación. ISSN: 1887-6587 Depósito legal: BA-178-07 Diseño: Grupo HABITAT. Imprime: Imprenta MORENO. Montijo (Badajoz, España). Grupo HABITAT. Centro de Investigación La Orden-Valdesequera. Apartado de Correos 22 (P.O. Box. 22) 06080 BADAJOZ (España). Secretaria General de Empleo, Actividad Empresarial e Innovación Tecnológica. Consejería de Empleo, Empresa e Innovación. FOLIA BOTANICA EXTREMADURENSIS Vol. 7 Julio 2013 Coordinación: Francisco Mª Vázquez Pardo Secretaría: Francisco Márquez García Equipo de edición: David García, María José Guerra, José Blanco y Francisco Márquez Equipo de redacción y revisión de textos: José Blanco Salas David García Alonso Francisco Márquez García José Luis Pérez Chiscano Carlos Pinto Gomes Francisco Mª Vázquez Pardo Ilustración de portada: Narcissus bulbocodium L. subsp. bulbocodium var. obesus (Salib.) Baker Edita: Consejería de Empleo, Empresa e Innovación. ISSN: 1887-6587 Depósito legal: BA-178-07 Diseño: Grupo HABITAT. Imprime: Imprenta MORENO. Montijo (Badajoz, España). Grupo HABITAT. -

Narcissus Species and Wild Hybrids
4- Narcissus Species and Wild Hybrids The following covers the wild species the system used throughout this Hand- and wild hybrids. They are grouped in book. Div. 10 of the Classified List and Inter- The genus Narcissus of the family national Register of Daffodil Names A maryilidaceae comprises 25 species in (1965) the present listing. Some authors list 30 Interest in growing Narcissus spans sev- or more species. These plants are na- eral hundred years and continues un- tives of the Mediterranean region, some abated to the present day. Despite the extending into central Europe. N. ta- wide popularity and interest in this zetta reportedly extends to China and plant in gardens, no modern botanical Japan. Assertions of the occurrence of treatment of the entire genus exists. tazeita in the Orient and the nebulous Much more collecting to bring together story of the origin of the Chinese Sacred new germ plasm for breeding purposes Lilies need more concentrated study be- and to learn more about the plants in fore acceptance. the wild is needed. With this in mind, I Narcissus is of particular interest to visited areas in Spain, Portugal, France, gardeners because most of the species and Italy in 1957 in part to collect wild may be hybridized with relative ease. Al- Narcissus. One of the interesting re- lowing for polyploidy, the resulting hy- sults of this trip was, for example, that brids will be at least partially fertile. It N. pseudo-norcissus subsp. tortuosus is possible to obtain an almost endless was found in the wild for the first time. -
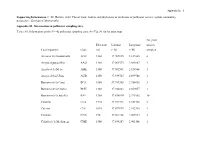
Supporting Information. C. M. Herrera. 2020. Flower Traits, Habitat, and Phylogeny As Predictors of Pollinator Service: a Plant Community Perspective
Appendix S1 – 1 Supporting Information. C. M. Herrera. 2020. Flower traits, habitat, and phylogeny as predictors of pollinator service: a plant community perspective. Ecological Monographs. Appendix S1. Information on pollinator sampling sites. TABLE S1. Information on the N = 42 pollinator sampling sites. See Fig. S1 for location map. No. plant Elevation Latitude Longitude species Local toponym Code (m) (º N) (º W) sampled Arenales del Guadalentín AGU 1360 37.909835 2.837405 4 Arroyo Aguaderillos AAG 1180 37.961573 2.883887 1 Arroyo de la Mesa AME 1100 37.902591 2.928966 1 Arroyo de los Ubios AUB 1250 37.939725 2.899766 1 Barranco de la Canal BCA 1580 37.789385 2.956438 3 Barranco de la Charca BCH 1360 37.942843 2.859057 1 Barranco de la Juanfría BJU 1380 37.836814 2.975302 10 Calarilla CLA 1710 37.944316 2.851708 2 Calerón CLE 1075 37.897973 2.942710 1 Cantalar CAN 770 37.964158 2.907974 3 Cañada de la Medianega CME 1580 37.894287 2.903146 1 Appendix S1 – 2 Cañada Pajarera CPA 1640 37.927456 2.805615 1 Collado del Calvario CCA 1420 37.950202 2.885484 1 Collado Zamora CZA 1420 37.839329 2.999615 2 Coto del Valle CVA 850 37.930542 2.928517 7 Cuesta del Bazar CBA 1330 37.906523 2.907470 1 Cuevas Bermejas CBE 1160 37.966959 2.851117 5 Fuente Bermejo FBE 1550 37.932268 2.840672 14 Fuente la Reina FRE 1460 37.941698 2.833632 2 Guadalentín GUA 1310 37.900404 2.836944 8 Hoyos de Muñoz HMU 1110 37.984831 2.876597 3 La Cabrilla LCA 1600 37.932364 2.780637 5 La Fresnedilla LFR 1080 37.980387 2.855875 2 La Mesa LME 1610 37.889823 2.913788 2 Las