Mitochondrial Genome Diversity in the Central Siberian Plateau with Particular
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Climate Change Among Nomadic and Settled Tungus of Siberia: Continuity
260 LAVRILLIER tundra and other geographical zones, pest outbursts over systems, 3) detection of correlation between the para- vast areas, and poor yields of forage resources; 4) the meters of climatic, hydrological, geocryologic and geo- decrease of some animal populations because of their morphologic events, on the one hand, and the intensity habitat reduction, adverse changes in the composition of negative processes in biological components of the of ichthyofauna, expansion of several animal and plant systems, on the other hand. This approach might allow species especially in the temperate and cold geographical forecasting some future adverse events and mitigating or zones of the northern hemisphere. The bulk of evidence eliminating their consequences. suggests that the present-day situation can be called a geoecological shock in the biosphere (in accordance References with the understanding of this term by V.I. Vernadsky Baskin, L.M. 2009. North reindeer. Moscow: KMK comradeship (Vernadsky 1998: 14–19). of scientific editions. Gorshkov, S.P. 2004.Global warming impact on nature, economy and society in central Siberia. In: Russian national work- Adaptation to the consequences of climate warming shop on research related to the international human dimen- as the major present-day task sions programme on global environmental change. Zvenig- Our civilization is not able to stop global warming in orod: Press of Russian Agricultural Academia (Proceedings 10–12): 194–203. the forthcoming decades. Therefore the problem of Mochalova, O.I. 2003. Climatic conditions and landscape adaptation associated with climate change may be more changes in the Angara-Yenisei Region. In: Climate, perma- urgent than IPCC ideas and conclusions suggest. -

The Fluvial Geochemistry of the Rivers of Eastern Siberia: I. Tributaries Of
Geochimica et Cosmochimica Acta, Vol. 62, No. 10, pp. 1657–1676, 1998 Copyright © 1998 Elsevier Science Ltd Pergamon Printed in the USA. All rights reserved 0016-7037/98 $19.00 1 .00 PII S0016-7037(98)00107-0 The fluvial geochemistry of the rivers of Eastern Siberia: I. Tributaries of the Lena River draining the sedimentary platform of the Siberian Craton 1, 1 2 1 YOUNGSOOK HUH, *MAI-YIN TSOI, ALEXANDR ZAITSEV, and JOHN M. EDMONd 1Department of Earth, Atmospheric and Planetary Sciences, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA 2Laboratory of Erosion and Fluvial Processes, Department of Geography, Moscow State University, Moscow, Russia (Received June 11, 1997; accepted in revised form February 12, 1998) ABSTRACT—The response of continental weathering rates to changing climate and atmospheric PCO2 is of considerable importance both to the interpretation of the geological sedimentary record and to predictions of the effects of future anthropogenic influences. While comprehensive work on the controlling mechanisms of contemporary chemical and mechanical weathering has been carried out in the tropics and, to a lesser extent, in the strongly perturbed northern temperate latitudes, very little is known about the peri-glacial environments in the subarctic and arctic. Thus, the effects of climate, essentially temperature and runoff, on the rates of atmospheric CO2 consumption by weathering are not well quantified at this climatic extreme. To remedy this lack a comprehensive survey has been carried out of the geochemistry of the large rivers of Eastern Siberia, the Lena, Yana, Indigirka, Kolyma, Anadyr, and numerous lesser streams which drain a pristine, high-latitude region that has not experienced the pervasive effects of glaciation and subsequent anthropogenic impacts common to western Eurasia and North America. -

Place Names: an Analysis of Published Materials
DOCUMENT RESUME ED 319 675 SO 020 925 AUTHOR Anderson, Paul S. TITLE Seeking a Core of Wo' -'d Regional Geography Place Names: An Analysis of Published Materials. PUB DATE 14 Oct 89 NOTE 18p.; Paper presentel at the Annual Meeting of the National Council for Geographic Education (Hershey, PA, October 11-14, 1989). Updated April 1990. PUB TYPE Speeches/Conference Papers (150) -- Reference Materials - Geographic Materials (133) -- Information Analyses (070) EDRS PRICE MF01/PC01 Plus Postage. DESCRIPTORS Elementary Secondary Education; *Geographic Location; *Geography Instruction; *Minimum Competencies; *Physical Geography IDENTIFIERS Place Names ABSTRACT Knowing place names is not the essence of geography, but some knowledge of names of geographical locations is widely considered to be basic information. Whether used in general cultural literacy, lighthearted Trivial Pursuit, educational sixth grade social studies, or serious debates on world events, place names and their locations are assumed to be known. At the college level of world regional geography courses, five books with lists of place names are in print by geographers: Fuson; MacKinnon; Pontius and Woodward; DiLisio; and Stoltman. Those five sources plus place name lists by P.S. Anderson and from Hirsch reveal similarities and diversities in their content. A core list of place names is presented with several cross-classifications by region, type of geographic feature, and grade level of students. The results reveal a logical progression of complexity that could assist geography educators to increase student learning and avoid duplication of efforts. There will never be complete agreement about any listing of the core geographical place names, but the presented lists are intended to stimulate discussion along constructive avenues. -

Structural Components of the Shamanic Rite of Tungusic Peoples in the Far East of Russia (On the Problem of Musical and Stylistic Specificity)
Journal of Siberian Federal University. Humanities & Social Sciences 6 (2016 9) 1410-1418 ~ ~ ~ УДК 39+781.7 Structural Components of the Shamanic Rite of Tungusic Peoples in the Far East of Russia (on the Problem of Musical and Stylistic Specificity) Svetlana V. Mezentseva* Khabarovsk State Institute of Culture 112 Krasnorechenskaia Str., Khabarovsk, 680045, Russia Received 08.02.2016, received in revised form 12.04.2016, accepted 07.05.2016 The subject of the work is a shamanic rite of the Tungusic peoples of the Far East of Russia. The purpose of the work is to highlight the main structural components of the shamanic rite in terms of musical and stylistic content, and provide a number of unique materials collected by the author during the field work. The paper analyzes the structure and musical and stylistic features of the elements of Nanai, Udege, Oroch, Ulch shamanistic rituals, and presents the transcriptions and analysis of the audio recording of the ritual of the known Nanai shaman Gara Kisovna Geiker. Keywords: ethnos, ethnic culture, music, shaman, shamanic rite, shamanistic ritual. DOI: 10.17516/1997-1370-2016-9-6-1410-1418. Research area: art history. Among the structural components of the The shamanistic curing rituals, which are musical and stylistic complex of a shamanic the subject of this research, were mostly popular pantheon, researchers distinguish shamanic among all Tungusic peoples. Curing shamanistic mythopoetical genres and special shamanistic rituals usually consisted of several basic parts: rituals. The latter had the following key functions: 1. Shaman’s calling spirit helpers - Curing of a patient for numerous ailments; 2. -

Climate Warming Impacts on Distributions of Scots Pine (Pinus Sylvestris L.) Seed Zones and Seed Mass Across Russia in the 21St Century
Article Climate Warming Impacts on Distributions of Scots Pine (Pinus sylvestris L.) Seed Zones and Seed Mass across Russia in the 21st Century Elena I. Parfenova 1, Nina A. Kuzmina 2, Sergey R. Kuzmin 2 and Nadezhda M. Tchebakova 1,* 1 Laboratory of Forest Monitoring, Sukachev Institute of Forest FRC KSC SB RAS, 660036 Krasnoyarsk, Russia; [email protected] 2 Laboratory of Forest Genetics and Breeding, Sukachev Institute of Forest FRC KSC SB RAS, 660036 Krasnoyarsk, Russia; [email protected] (N.A.K.); [email protected] (S.R.K.) * Correspondence: [email protected] Abstract: Research highlights: We investigated bioclimatic relationships between Scots pine seed mass and seed zones/climatypes across its range in Russia using extensive published data to predict seed zones and seed mass distributions in a changing climate and to reveal ecological and genetic components in the seed mass variation using our 40-year common garden trial data. Introduction: seed productivity issues of the major Siberian conifers in Asian Russia become especially relevant nowadays in order to compensate for significant forest losses due to various disturbances during the 20th and current centuries. Our goals were to construct bioclimatic models that predict the seed mass of major Siberian conifers (Scots pine, one of the major Siberian conifers) in a warming climate during the current century. Methods: Multi-year seed mass data were derived from the literature and were collected during field work. Climate data (January and July data and annual precipitation) were Citation: Parfenova, E.I.; Kuzmina, derived from published reference books on climate and climatic websites. -

Mapping Russian Census 2002
CENSUS ATLAS OF RUSSIA: FERTILITY An NCEEER Working Paper by Timothy Heleniak American Geographical Society National Council for Eurasian and East European Research 1828 L Street NW Suite 1200 Washington, DC 20036 [email protected] http://www.nceeer.org/ TITLE VIII PROGRAM Project Information* Principal Investigator: Timothy Heleniak NCEEER Contract Number: 828-06 Date: August 29, 2014 Copyright Information Individual researchers retain the copyright on their work products derived from research funded through a contract or grant from the National Council for Eurasian and East European Research (NCEEER). However, the NCEEER and the United States Government have the right to duplicate and disseminate, in written and electronic form, reports submitted to NCEEER to fulfill Contract or Grant Agreements either (a) for NCEEER’s own internal use, or (b) for use by the United States Government, and as follows: (1) for further dissemination to domestic, international, and foreign governments, entities and/or individuals to serve official United States Government purposes or (2) for dissemination in accordance with the Freedom of Information Act or other law or policy of the United States Government granting the public access to documents held by the United States Government. Neither NCEEER nor the United States Government nor any recipient of this Report may use it for commercial sale. * The work leading to this report was supported in part by contract or grant funds provided by the National Council for Eurasian and East European Research, funds which were made available by the U.S. Department of State under Title VIII (The Soviet-East European Research and Training Act of 1983, as amended). -
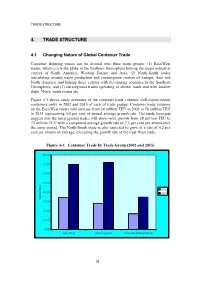
4. Trade Structure
TRADE STRUCTURE 4. TRADE STRUCTURE 4.1 Changing Nature of Global Container Trade Container shipping routes can be divided into three main groups: (1) East-West trades, which circle the globe in the Northern Hemisphere linking the major industrial centres of North America, Western Europe and Asia; (2) North-South trades articulating around major production and consumption centres of Europe, Asia and North America, and linking these centres with developing countries in the Southern Hemisphere; and (3) intraregional trades operating in shorter hauls and with smaller ships. North–south routes are Figure 4-1 shows study estimates of the container trade volumes (full export/import containers only) in 2002 and 2015 of each of trade groups. Container trade volumes on the East-West routes will increase from 34 million TEU in 2002 to 70 million TEU in 2015 representing 5.8 per cent of annual average growth rate. The study forecasts suggest that the intraregional trades will show solid growth from 28 million TEU to 72 million TEU with a compound average growth rate of 7.5 per cent per annum over the same period. The North-South trade is also expected to grow at a rate of 6.2 per cent per annum on average, exceeding the growth rate of the East-West trade. Figure 4-1: Container Trade by Trade Group (2002 and 2015) 80.00 70.00 60.00 50.00 2002 40.00 2015 Million TEU 30.00 20.00 10.00 0.00 East-West Intra-Regional North-South/South-South 32 TRADE STRUCTURE 4.2 Asia - North America The biggest deep sea liner route is the trans-Pacific trade between Asia and North America, representing 14.5 million TEU in 2002, equivalent to 43 per cent of the total East-West trade and 19 per cent of the world total. -

Changes in the Arctic: Background and Issues for Congress
Changes in the Arctic: Background and Issues for Congress Updated May 22, 2020 Congressional Research Service https://crsreports.congress.gov R41153 Changes in the Arctic: Background and Issues for Congress Summary The diminishment of Arctic sea ice has led to increased human activities in the Arctic, and has heightened interest in, and concerns about, the region’s future. The United States, by virtue of Alaska, is an Arctic country and has substantial interests in the region. The seven other Arctic states are Canada, Iceland, Norway, Sweden, Finland, Denmark (by virtue of Greenland), and Russia. The Arctic Research and Policy Act (ARPA) of 1984 (Title I of P.L. 98-373 of July 31, 1984) “provide[s] for a comprehensive national policy dealing with national research needs and objectives in the Arctic.” The National Science Foundation (NSF) is the lead federal agency for implementing Arctic research policy. Key U.S. policy documents relating to the Arctic include National Security Presidential Directive 66/Homeland Security Presidential Directive 25 (NSPD 66/HSPD 25) of January 9, 2009; the National Strategy for the Arctic Region of May 10, 2013; the January 30, 2014, implementation plan for the 2013 national strategy; and Executive Order 13689 of January 21, 2015, on enhancing coordination of national efforts in the Arctic. The office of the U.S. Special Representative for the Arctic has been vacant since January 20, 2017. The Arctic Council, created in 1996, is the leading international forum for addressing issues relating to the Arctic. The United Nations Convention on the Law of the Sea (UNCLOS) sets forth a comprehensive regime of law and order in the world’s oceans, including the Arctic Ocean. -

Manchu Grammar (Gorelova).Pdf
HdO.Gorelova.7.vw.L 25-04-2002 15:50 Pagina 1 MANCHU GRAMMAR HdO.Gorelova.7.vw.L 25-04-2002 15:50 Pagina 2 HANDBOOK OF ORIENTAL STUDIES HANDBUCH DER ORIENTALISTIK SECTION EIGHT CENTRAL ASIA edited by LILIYA M. GORELOVA VOLUME SEVEN MANCHU GRAMMAR HdO.Gorelova.7.vw.L 25-04-2002 15:50 Pagina 3 MANCHU GRAMMAR EDITED BY LILIYA M. GORELOVA BRILL LEIDEN • BOSTON • KÖLN 2002 HdO.Gorelova.7.vw.L 25-04-2002 15:50 Pagina 4 This book is printed on acid-free paper Die Deutsche Bibliothek – CIP-Einheitsaufnahme Gorelova, Liliya M.: Manchu Grammar / ed. by Liliya M. Gorelova. – Leiden ; Boston ; Köln : Brill, 2002 (Handbook of oriental studies : Sect.. 8, Central Asia ; 7) ISBN 90–04–12307–5 Library of Congress Cataloging-in-Publication Data Gorelova, Liliya M. Manchu grammar / Liliya M. Gorelova p. cm. — (Handbook of Oriental Studies. Section eight. Central Asia ; vol.7) Includes bibliographical references and index. ISBN 9004123075 (alk. paper) 1. Manchu language—Grammar. I. Gorelova, Liliya M. II. Handbuch der Orientalis tik. Achte Abteilung, Handbook of Uralic studies ; vol.7 PL473 .M36 2002 494’.1—dc21 2001022205 ISSN 0169-8524 ISBN 90 04 12307 5 © Copyright 2002 by Koninklijke Brill NV, Leiden, The Netherlands All rights reserved. No part of this publication may be reproduced, translated, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without prior written permission from the publisher. Authorization to photocopy items for internal or personal use is granted by E.J. Brill provided that the appropriate fees are paid directly to The Copyright Clearance Center, 222 Rosewood Drive, Suite 910 Danvers MA 01923, USA. -

Historical Sciences 85
Historical sciences 85 Short Report THE CALENDAR VOCABULARY AS A STUDY relations of the Samoyeds and Yenisseys it may be SOURCE OF ANCIENT CULTURAL- supposed the following peculiarities of the origin of LINGUISTIC CONTACTS OF SIBERIAN the names for conceptions “spring”, “winter” in the INDIGENOUS PEOPLES (TO THE PROBLEM Samoyed (Selkup) and the Yenissey (Ket) languages: OF THE ORIGIN OF THE WORDS “SPRING”, 1) The words “winter”, “spring” had been bor- “WINTER»IN THE SELKUP AND THE KET rowed by native speakers from each other during the LANGUAGES) period of the existence of Samoyed and Yenissey lin- Kolesnikova V. guistic communities. [email protected] 2) The words “winter”, “spring” had been bor- rowed by native speakers from each other after the Different ethnic groups had been living, mov- disintegration of indicated linguistic communities (or ing, assimilating on the territory of Western Siberia one of them). for a long time. Economic and cultural contacts be- 3) The words “winter”, “spring” have been tween representatives of these groups had been arising borrowed by native speakers not from each other, but during many years. The relations and the two-way from another source. influence of two peoples – the Samoyeds and Yenis- It is known that there are many words in seys (ancestors of modern Selkups and Kets) - is one Selkup and Ket borrowed from other, unrelated lan- example of such contacts. Samoyed-yenissey lexical guages. For example, words from Russian and Turkic equivalents demonstrated by some researchers may be have been fixed in the Selkup language. A whole given as the results of their relationship [1]. -

Second Report Submitted by the Russian Federation Pursuant to The
ACFC/SR/II(2005)003 SECOND REPORT SUBMITTED BY THE RUSSIAN FEDERATION PURSUANT TO ARTICLE 25, PARAGRAPH 2 OF THE FRAMEWORK CONVENTION FOR THE PROTECTION OF NATIONAL MINORITIES (Received on 26 April 2005) MINISTRY OF REGIONAL DEVELOPMENT OF THE RUSSIAN FEDERATION REPORT OF THE RUSSIAN FEDERATION ON THE IMPLEMENTATION OF PROVISIONS OF THE FRAMEWORK CONVENTION FOR THE PROTECTION OF NATIONAL MINORITIES Report of the Russian Federation on the progress of the second cycle of monitoring in accordance with Article 25 of the Framework Convention for the Protection of National Minorities MOSCOW, 2005 2 Table of contents PREAMBLE ..............................................................................................................................4 1. Introduction........................................................................................................................4 2. The legislation of the Russian Federation for the protection of national minorities rights5 3. Major lines of implementation of the law of the Russian Federation and the Framework Convention for the Protection of National Minorities .............................................................15 3.1. National territorial subdivisions...................................................................................15 3.2 Public associations – national cultural autonomies and national public organizations17 3.3 National minorities in the system of federal government............................................18 3.4 Development of Ethnic Communities’ National -

Ancient Fennoscandian Genomes Reveal Origin and Spread of Siberian Ancestry in Europe
ARTICLE DOI: 10.1038/s41467-018-07483-5 OPEN Ancient Fennoscandian genomes reveal origin and spread of Siberian ancestry in Europe Thiseas C. Lamnidis1, Kerttu Majander1,2,3, Choongwon Jeong1,4, Elina Salmela 1,3, Anna Wessman5, Vyacheslav Moiseyev6, Valery Khartanovich6, Oleg Balanovsky7,8,9, Matthias Ongyerth10, Antje Weihmann10, Antti Sajantila11, Janet Kelso 10, Svante Pääbo10, Päivi Onkamo3,12, Wolfgang Haak1, Johannes Krause 1 & Stephan Schiffels 1 1234567890():,; European population history has been shaped by migrations of people, and their subsequent admixture. Recently, ancient DNA has brought new insights into European migration events linked to the advent of agriculture, and possibly to the spread of Indo-European languages. However, little is known about the ancient population history of north-eastern Europe, in particular about populations speaking Uralic languages, such as Finns and Saami. Here we analyse ancient genomic data from 11 individuals from Finland and north-western Russia. We show that the genetic makeup of northern Europe was shaped by migrations from Siberia that began at least 3500 years ago. This Siberian ancestry was subsequently admixed into many modern populations in the region, particularly into populations speaking Uralic languages today. Additionally, we show that ancestors of modern Saami inhabited a larger territory during the Iron Age, which adds to the historical and linguistic information about the population history of Finland. 1 Department of Archaeogenetics, Max Planck Institute for the Science of Human History, 07745 Jena, Germany. 2 Institute for Archaeological Sciences, Archaeo- and Palaeogenetics, University of Tübingen, 72070 Tübingen, Germany. 3 Department of Biosciences, University of Helsinki, PL 56 (Viikinkaari 9), 00014 Helsinki, Finland.