A Mitochondrial Analysis Reveals Distinct Founder Effect Signatures in Canarian and Balearic Goats
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Rankings Municipality of Artenara
9/29/2021 Maps, analysis and statistics about the resident population Demographic balance, population and familiy trends, age classes and average age, civil status and foreigners Skip Navigation Links SPAGNA / CANARIAS / Province of LAS PALMAS / ARTENARA Powered by Page 1 L'azienda Contatti Login Urbistat on Linkedin Adminstat logo DEMOGRAPHY ECONOMY RANKINGS SEARCH SPAGNA Municipalities Powered by Page 2 AGAETE Stroll up beside >> L'azienda Contatti Login Urbistat on Linkedin LA OLIVA AdminstatAGÜIMES logo DEMOGRAPHY ECONOMY RANKINGS SEARCH LAS PALMAS ANTIGUA SPAGNA DE GRAN ARRECIFE CANARIA ARTENARA MOGÁN ARUCAS MOYA BETANCURIA PÁJARA FIRGAS PUERTO DEL GÁLDAR ROSARIO HARÍA SAN BARTOLOMÉ INGENIO SAN LA ALDEA DE BARTOLOMÉ SAN NICOLÁS DE TIRAJANA SANTA BRÍGIDA SANTA LUCÍA DE TIRAJANA STA. MARÍA DE GUÍA DE G.C. TEGUISE TEJEDA TELDE TEROR TÍAS TINAJO TUINEJE VALLESECO VALSEQUILLO DE GRAN CANARIA VEGA DE SAN MATEO YAIZA Provinces Powered by Page 3 LAS PALMAS SANTA CRUZ DE L'azienda Contatti Login Urbistat on Linkedin TENERIFE Adminstat logo DEMOGRAPHY ECONOMY RANKINGS SEARCH SPAGNA Regions ANDALUCÍA CEUTA ARAGÓN COMUNIDAD DE MADRID CANARIAS COMUNIDAD CANTABRIA FORAL DE CASTILLA - LA NAVARRA MANCHA COMUNITAT CASTILLA Y VALENCIANA LEÓN EXTREMADURA CATALUÑA GALICIA ILLES BALEARS LA RIOJA MELILLA PAÍS VASCO PRINCIPADO DE ASTURIAS REGIÓN DE MURCIA Municipality of Artenara Territorial extension of Municipality of ARTENARA and related population density, population per gender and number of households, average age and incidence of foreigners TERRITORY -

08.MAAL 8De15.Pdf
226 M. A. Arnedo and C. Ribera Table V. Intraspecific spination variability o/Dysdera paucispinosa Proximal Med.-proximal Medial-distal Distal Tibia 3 dorsal 0-1.0.0 0 0 1.0.0 Tibia 4 dorsal 0.0.0-1 0 0 0 Tibia 3 ventral 0.0-1.0 0 0 0 Tibia 4 ventral 0.1-2.0 0 0 0 Number of rows Number of spines Femur 3 dorsal 0 — Femur 4 dorsal " 0 from middle area to the front; small scale or ridge present at both sides of S attachment. S arms are shorter than DA; straight; tips dorsally projected; neck as wide as arms. TB usual shape. ALS (Fig. 19A) with pyriform gland spigot in polar position; remaining pyriform spigots more external than major ampulate gland spigot, arranged in one row; 4 +1 pyriform gland spigots; PMS and PLS (Fig. 19B-C) with fewer than 10 aciniform gland spigots. Intraspecific variation. P sometimes toothed along its upper margin. Spination variability in Table 5. Distribution. Gran Cañarían endemic. Relatively rare species collected from pine forest-related habitats. Comments. After examination of type material of D. paucispinosa and D. tamadabaensis no non-sex-related character to distinguish the two species was found. Therefore, the second species is considered to be the male of the first and hence a junior synonym. Dysdera tibicena sp. n. (Figs. 20A-H, 21A-D, 22A-C) Holotypemale. 14/2/96, MA. Arnedo, B. Emerson, R. Fragoso, C. Juan & P. Oromi leg.; num. 3049, stored at UB. Type locality. Pinar de Tamadaba, Agaete, Gran Canaria, Canary Islands. -

220 220 Horarios
Horarios 220 Las Palmas de G.C. - Artenara HORAS / HOURS 07 08 09 10 11 12 13 14 15 16 17 18 19 21 minutos/minutes: Lunes a viernes / From 00 00 00 00 00 00 00 15 00 00 00 00 00 30 monday to friday / Von Montag bis Freitag Sábado / Saturday / 00 00 00 00 00 00 00 00 15 00 00 30 Samstag Festivos / Bank holiday / 00 00 00 00 00 15 00 15 00 00 30 Feiertagen 15 Desde Teror.Hasta Valsendero. Desde Teror.Hasta Tejeda.Por Artenara. Desde Teror.Por Juncalillo. Desde Teror.Hasta Lanzarote. Recorrido. Estación San Telmo (andén 18) , Subida de Mata , Las Rehoyas , Cruz de Piedra , Las Palmas de G.C. (CC La Ballena) , La Paterna , Cruce Los Tarahales , Los Tarahales , Colegio Arenas , CC Siete Palmas , Cruce de Almatriche , Lomo Cuesta Blanca , Hoya Andrea , Cruce Las Torres , Barranco de Tamaraceite , Iglesia de Tamaraceite , Recta de Tamaraceite , Tamaraceite , La Suerte , Pileta , Rotonda Ciudad del Campo , Toscón Bajo , Toscón Alto , Casa Camineros , El Chaparral , Barranco Zapatero , Parada del Francés , Casa Vieja , Miraflor , Llano de Arevalo , Vuelta de Los Alambres , Puente Molino , Entrada Roque Nublo , Fábrica de Donuts , El Hoyo , Casa Juanero , Teror , Camino al Rincón , San Matías , Avd. Cabildo , Avd. Cabildo , Avd. Cabildo , El Rincón , Las Rosadas , Entrada Las Rosadas , La Granja , Entrada Hipodromo , Cruce de la Laguna , Mirador de Zamora , Valleseco , Gasolinera de Valleseco , Las Cuevas , El Balcón , Autoservicios Mesas , Lanzarote , Cruce de Tejeda , Cueva Corcho , Retamilla , Montañon Negro , El Tablado , Juncalillo , El Tablado , Entrada Juncalillo , Las Arbejas , Cueva de Los Gatos , Lomo Cuchara , Artenara . -

Risco Caído and the Sacred Mountains of Gran Canaria Cultural Landscape
Additional information requested by ICOMOS regarding the nomination of the Risco Caído and the Sacred Mountains of Gran Canaria Cultural Landscape for Inscription on the World Heritage List 2018 November 2018 1 Index This report includes the additional information requested by ICOMOS in its letter of the 8th October 2018 concerning the nomination process of Risco Caido and Sacred Mountains of Gran Canaria Cultural Landscape. It includes the information requested, along with the pertinent comments on each point. 1. Description of de property p. 3 2. Factors affecting the property p. 54 3. Boundaries and the buffer zone p. 59 4. Protection p. 68 5. Conservation p. 79 6. Management p. 87 7. Involvement of the local communities p. 93 2 1 Description of the property ICOMOS would be pleased if the State Party could provide a more accurate overview of the current state of archaeological research in the Canary Islands in order to better understand Gran Canaria's place in the history of the archipelago. The inventory project begun at the initiative of Werner Pichler which mentions the engravings of the north of Fuerteventura with 2866 individual figures and the work briefly mentioned in the nomination dossier of several researchers from the Universidad de La Laguna, on the island of Tenerife, and the Universidad de Las Palmas, on the island of Gran Canaria could assist in this task. Table 2.a.llists all the attributes and components of the cultural landscape of Risco Caldo and its buffer zone (p. 34). However, only part of the sites are described in the nomination dossier (p. -

Ruta Jacobea Santiago De Tunte - Santiago De Gáldar 84 Km
RUTA JACOBEA SANTIAGO DE TUNTE - SANTIAGO DE GÁLDAR 84 km. CICLOTURISMO BICYCLE TOURING Santiago de Gáldar FAHRRADTOURISMUS Agaete Las Palmas CICLOTURISMO de Gran Canaria San Nicolás Tejeda RUTA JACOBEA de Tolentino Ayacata SANTIAGO DE TUNTE - SANTIAGO DE GÁLDAR Santiago de Tunte Tunte Agüimes Puerto de Mogán Bahía Feliz San Agustín Arguineguín Playa del Inglés Meloneras Maspalomas Gran Canaria es uno de los mejores lugares del mundo para practicar ciclismo. Cuenta con una excelente infraestructura de carreteras, hospitales, hoteles y demás servicios que harán la estancia cicloturista inolvidable. Gran Canaria is one of the best spots in the world to go cycling. It has an excellent infrastructure of roads, hospitals, hotels and other services that will make your biking stay unforgettable. Gran Canaria: Eines der beliebtesten Reiseziele für Radsportler weltweit! Eine ausgezeichnete Infrastruktur mit Straßen, Krankenhäusern, Hotels und allen erdenklichen Serviceleistungen lassen Ihren Radsporturlaub unvergesslich werden. Patronato de Turismo de Gran Canaria Triana, 93 Gran Canaria è uno dei migliori posti al mondo per il ciclismo. Possiede buone 35002 Las Palmas de Gran Canaria infrastrutture stradali, ospedaliere, alberghiere e altri servizi che renderanno Tel: (+34) 928 219 600 indimenticabile il soggiorno cicloturistico. Fax: (+34) 928 219 601 Ruta Jacobea: Santiago de Tunte - “The way of St James” route: Santiago Santiago de Gáldar. 84 km. de Tunte - Santiago de Gáldar. 84 km. Salimos de la plaza de la iglesia donde está la Puerta Santa, We leave the church square in front of the church main en SANTIAGO DE TUNTE - SAN BARTOLOMÉ DE TIRAJANA entrance, in SANTIAGO DE TUNTE or SAN BARTOLOMÉ DE por la GC-60 con una subida de 6 km. -

Artenara Area
Description of other walks available in the Artenara area The following 4 walks are all excellent walks that are not included in my two Don’t Leave Gran Canaria walking guides “Don’t leave Gran Canaria without seeing it”. You will walk past many occupied cave houses and villages, all on well maintained paths, often Without Seeing signposted with excellent scenery. These 4 walks are available in single page booklet style and are available from Octavio in the Artenara tourist office. Walk 1:- A 2 hour 15 minute circular walk to Las Arbejas, then onto a wonderful viewing area above Artenara. Returning via many cave houses, a cave Artenara church and museum. Ascent and Descent approx 275m. Walk 2:- A 2 hour 15 minute circular walk around the Tamadaba forest. This walk can be extended to make a 4 hour plus walk back to Artenara. Ascent and Descent approx 300m Walk 3:- A 2 hour 20 minute circular walk that heads towards the Tejeda valley on an excellent Camino Real, returning to Artenara on a delightful forest path. Ascent and Descent approx 300m Walk 4:- A 3 hour 15 minute circular walk with excellent views around several villages of occupied cave houses. Ascent and Descent approx 400 metres. There are many other excellent half day and full day walks, in this area, in both Volume 1 and Volume 2 of my walking guides ”Don’t leave Gran Canaria without seeing it”. All the walks in both of my guide books have full descriptions, maps and downloadable GPX tracks and waypoints. -

EMSR379: Forest Fire in Artenara, Spain EMSR381: Forest Fire in Cazadores, Spain EMSR382: Forest Fire in Valleseco, Gran Canaria, Spain - Information Bulletin 120
EMSR379: Forest fire in Artenara, Spain EMSR381: Forest fire in Cazadores, Spain EMSR382: Forest fire in Valleseco, Gran Canaria, Spain - Information Bulletin 120 1 / 9 Gran Canaria, one of Spain’s Canary Islands, was struck by forest fires in two locations recently – in Artenara on 10 August, Cazadores on 12 August and Valleseco on 17 August (in the same area as the Artenara fire). The Copernicus EMS Rapid Mapping module was activated for each of these forest fires and tasked with providing delineation and grading products. Media reports state that these forest fires were caused by human actions but were aggravated by drought conditions and strong winds. The situation was unfortunately predictable due to the prolonged heatwaves during summer 2019, which have affected the territory of Gran Canaria. According to media reports, around 10,000 people at a single point in time have been evacuated as a result of these forest fires, while the number of first responders engaged rose above 1,000, supported by 16 aircraft. On 13 August the EMS Rapid Mapping module was activated by the Operational Coordination Center of the Directorate General of Civil Protection and Emergencies in Spain following a request from the Government of the Canary Islands for the forest fire in Artenara. The next day, one grading product was produced using optical imagery acquired from the SPOT6 satellite to get an overview of the forest fire situation in the area. This grading product showed that 1,163.3 ha (11.63 km2) of land, 308 residential buildings and 2,453 people were affected by the fire. -

Endemics Versus Newcomers: the Ladybird Beetle (Coleoptera: Coccinellidae) Fauna of Gran Canaria
insects Article Endemics Versus Newcomers: The Ladybird Beetle (Coleoptera: Coccinellidae) Fauna of Gran Canaria Jerzy Romanowski 1,* , Piotr Ceryngier 1 , Jaroslav Vetrovec˘ 2, Marta Piotrowska 3 and Karol Szawaryn 4 1 Institute of Biological Sciences, Cardinal Stefan Wyszy´nskiUniversity, Wóycickiego 1/3, 01-938 Warsaw, Poland; [email protected] 2 Buzulucká, 1105 Hradec Králové, Czech Republic; [email protected] 3 Faculty of Biology and Environmental Sciences UKSW, ul. Wóycickiego 1/3, PL-01-938 Warsaw, Poland; [email protected] 4 Museum and Institute of Zoology, Polish Academy of Sciences, Wilcza 64, 00-679 Warsaw, Poland; [email protected] * Correspondence: [email protected] Received: 18 August 2020; Accepted: 14 September 2020; Published: 18 September 2020 Simple Summary: Many plants and animals that live in the Canary Islands belong to the so-called endemic species, i.e., they do not occur outside of this particular region. Several other species have a slightly wider geographical distribution, apart from the Canaries, which also includes some islands of the nearby archipelagos, such as Madeira or the Azores, or the northwestern periphery of Africa. Here, we call such species subendemics. However, the Canary Islands have recently been colonized by a substantial number of immigrants from more or less remote areas. In this paper, based on our field survey and previously published data, we analyzed the fauna of the ladybird beetles (Coccinellidae) of Gran Canaria, one of the central islands of the archipelago. Among 42 ladybird beetle species so far recorded on this island, 17 (40%) are endemics and subendemics, and 21 (50%) probably arrived in Gran Canaria relatively recently, i.e., in the 20th and 21st century. -

A Mayores Dependientes En Gran Canaria
PERFIL DE ATENCIÓN SOCIOSANITARIA A MAYORES DEPENDIENTES EN GRAN CANARIA Dra Pilar PérezPérez Responsable del Servicio de Admisión INSTITUTO-AS Cabildo de Gran Canaria Las Palmas de Gran Canaria, septiembre 2015 SERVICIO DE ADMISION DEFINICIÓN Y ORGANIZACIÓN Y DEL SERVICIO DE ADMISIÓN ü SERVICIO MULTIDISCIPLINAR DE CARÁCTER SOCIOSANITARIO ü DEPENDENCIA JERÁRQUICA DIRECTA DE LA GERENCIA DEL INSTITUTO DE ATENCION SOCIAL Y SOCIOSANITARA (IAS), CABILDO DE GRAN CANARIA ü INICIO DE ACTIVIDAD EN EL AÑO 2000 (PCAMD 2002) ü COMPOSICIÓN ACTUAL: • 2 ADMINISTRATIVOS • 3 TRABAJADORES SOCIALES • 1 DUE • 1 MÉDICO SERVICIO DE ADMISIÓN FUNCIÓN DEL SERVICIO DE ADMISIÓN GESTIÓN DEL ACCESO A LA RED DE RECURSOS SOCIOSANITARIOS DEPENDIENTES DEL IAS SERVICIOS ESPECIALIZADOS CENTROS RESIDENCIALES Y CENTROS DE ESTANCIA DIURNA AREAS DE ACTUACIÓN MAYORES Y DISCAPACIDAD (Sectores Discapacidad y Salud Mental) ÁMBITO INSULAR DE GRAN CANARIA SERVICIO DE ADMISIÓN FUNCIONES DEL SERVICIO DE ADMISIÓN ü Información, orientación y asesoramiento a usuarios, familiares, agentes sociosanitarios y público en general. Atención directa. ü Asesoría y apoyo técnico a otros agentes internos y externos. ü Coordinación entre los Centros gestionados por el Instituto AS y otros recursos del ámbito Socio-Sanitario. ü Gestión, tramitación y valoración de traslados de usuarios. ü Gestión, tramitación y valoración de casos de urgencia sociosanitaria. ü Gestión, tramitación y valoración de solicitudes de exención de pago. ü Valoración presencial de usuarios en domicilio y recursos sociosanitarios y sanitarios públicos y privados. ü Participación y preparación de Comisiones técnicas. ü Actualización y gestión de la base de datos de usuarios. ü Intervención en situaciones de crisis, urgencia y/o emergencia social. -
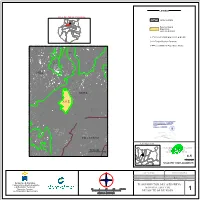
T:\Planesredcanariaenp\C
LEYENDA ISLA DE GRAN CANARIA Límite municipal SANTA MARIA MOYA ARUCAS GALDAR DE GUIA DE G. C. FIRGAS LAS PALMAS DE AGAETE GRAN CANARIA SITUACIÓN TEROR VALLESECO SANTA BRIGIDA ARTENARA Reserva Natural VEGA DE SAN MATEO TELDE VALSEQUILLO DE GRAN CANARIA Especial de TEJEDA SAN NICOLAS DE TOLENTINO Los Tilos de Moya INGENIO AGÜIMES MOGAN SAN BARTOLOME DE TIRAJANA SANTA LUCIA DE TIRAJANA C-3=Reserva Natural Especial de El Brezal C-12=Parque Rural de Doramas TRUJILLO C-4=Reserva Natural Especial de Azuaje Lomo De De Barranco Las carreras Abalos MA Casas de aguilar Moya Los LOMO Barranco Barranco De Palmitos Nocas EL LANCE Santa ines Los desaguaderos Barranco LOS C-12 Las PALO Los paredones Finca Barranco De EL San francisco Barranquillo Los gramales Paso de maria de los santos Vergara El palmito Nocas El palo Brezal Los paredones El penonal Morejones Finca la Las cuartas El calvario Vinculo Renta Los desaguaderos De solis Lomo MA De El granillar Palma Del Los El curato Zarzas El gamonal PLAT Los silillos Barranco Moya Torres De La suerte MOYA DEHESILLA LOS CASTILLEJOS HOYAS VIVAS El cercado Las Las Barranco Azuaje HOYA DE MA C-3 PEDRO Las vinagreras Pajarito El palmital bajo De Pico lomito EL PINILLO Carreteria Quintana EL BREZAL Lomo SOLAPONES MA La capellania MA Castillejo De MA La ladera LOS La penilla C-4 Cruz MA MA MA MA La Lomo HOYA De MA Barranquillo frio GUÍA De Barranco De Lomo PLAT LOS LLANETES Guzman DE MA MA MA El flande De La LOMO EL TRAPICHILLO Las canales El palmital alto La josefa MA Corona Llano de MATOS BRACAMONTE CRISTINA -

Documento De Pliegos Número De Expediente 93/2020 Publicado En La Plataforma De Contratación Del Sector Público El 10-06-2020 a Las 16:57 Horas
Documento de Pliegos Número de Expediente 93/2020 Publicado en la Plataforma de Contratación del Sector Público el 10-06-2020 a las 16:57 horas. Mejora del Alumbrado Público del Norte de Gran Canaria, financiado por el Plan de Cooperación del Cabildo de Gran Canaria (19.PCM.23.01). Valor estimado del contrato 396.852,25 EUR. Tipo de Contrato Obras Importe 424.631,91 EUR. Subtipo Instalación eléctrica Importe (sin impuestos) 396.852,25 EUR. Lugar de ejecución ES70 Canarias Proyecto a realizar en Plazo de Ejecución los municipios de Agaete, Artenara, Firgas, Gáldar, La 1 Mes(es) Aldea de San Nicolás y Valleseco, en la Isla de Gran Observaciones: 4 semanas todos los lotes, salvo el Canaria, Provincia de Las Palmas. Municipios de Agaete, de Gáldar 8 semanas Artenara, Firgas, Gáldar, La Aldea de San Nicolás y Valleseco. Clasificación CPV 45316000 - Trabajos de instalación de sistemas de alumbrado y señalización. Pliego Prescripciones Técnicas Pliego Cláusulas Administrativas ANEXO LOTE AGAETE.pdf ANEXO LOTE ARTENARA.pdf ANEXO LOTE FIRGAS.pdf ANEXO LOTE GALDAR.pdf ANEXO LOTE LA ALDEA.pdf ANEXO LOTE VALLESECO.pdf Proceso de Licitación Procedimiento Abierto simplificado Tramitación Ordinaria Presentación de la oferta Electrónica Detalle de la Licitación: https://contrataciondelestado.es/wps/poc?uri=deeplink:detalle_licitacion&idEvl=d0UiYC2nEbVvYnTkQN0%2FZA%3D%3D Nº de Lotes: 6 Se debe ofertar: A uno o varios lotes Número máximo de lotes que se puede adjudicar a un licitador: 6 Entidad Adjudicadora Presidencia de la Mancomunidad de Ayuntamientos -

Plan De Inversiones Para La Reactivación Económica De Gran
Programa de Inversiones y Nuevas Medidas Sociales para la reactivación económica y social de Gran Canaria 2 Programa de Inversiones para la reactivación económica y social de Gran Canaria #JuntosSaldremosDeEsta Consejería: Turismo Ubicación Descripción Importe San Bartolomé Proyecto embellecimiento GC-1 a la altura del 450.000,00 de Tirajana Veril (Consorcio de Maspalomas) San Bartolomé Aparcamiento en Playa del Águila (Consorcio 239.696,15 de Tirajana de Maspalomas) San Bartolomé Rehabilitación del Paseo Meloneras (Consorcio 500.000,00 de Tirajana de Maspalomas) San Bartolomé Alumbrado Público Playa del inglés Anexo I a El 170.000,00 de Tirajana Veril (Consorcio de Maspalomas) San Bartolomé Embellecimiento Pétreo Muro GC-500 Punto 80.000,00 de Tirajana Kilométrico 15.800 (Consorcio de Maspalomas) Las Palmas de Proyecto Las Canteras sombras digitales 80.000,00 Gran Canaria La Laja. Estructura contenedor acero inoxidable doble, accesible, lavamanos central, Las Palmas de con un kit de autosuficiencia mediante tanques 52.000,00 Gran Canaria integrados para agua limpia, residual y con energía solar, apto PMR Las Palmas de La Laja. Mejora del entorno de las piscinas de 161.364,55 Gran Canaria La Laja Las Palmas de Las Canteras. Rehabilitación de base náutica y 1.707.589,60 Gran Canaria balnearios en la puntilla Las Canteras. Mejoras de accesibilidad Las Palmas de universal. Itinerario asistencia baño en Las 1.048.586,85 Gran Canaria Canteras. Diversas actuaciones de mejoras en el término Mogán 1.350.000,00 municipal de Mogán Modificación e