Structure of the Full Kinetoplastids Mitoribosome and Insight on Its Large Subunit Maturation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Disability Classification System
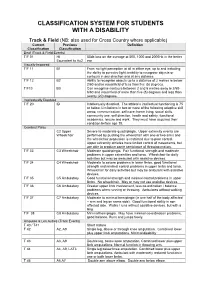
CLASSIFICATION SYSTEM FOR STUDENTS WITH A DISABILITY Track & Field (NB: also used for Cross Country where applicable) Current Previous Definition Classification Classification Deaf (Track & Field Events) T/F 01 HI 55db loss on the average at 500, 1000 and 2000Hz in the better Equivalent to Au2 ear Visually Impaired T/F 11 B1 From no light perception at all in either eye, up to and including the ability to perceive light; inability to recognise objects or contours in any direction and at any distance. T/F 12 B2 Ability to recognise objects up to a distance of 2 metres ie below 2/60 and/or visual field of less than five (5) degrees. T/F13 B3 Can recognise contours between 2 and 6 metres away ie 2/60- 6/60 and visual field of more than five (5) degrees and less than twenty (20) degrees. Intellectually Disabled T/F 20 ID Intellectually disabled. The athlete’s intellectual functioning is 75 or below. Limitations in two or more of the following adaptive skill areas; communication, self-care; home living, social skills, community use, self direction, health and safety, functional academics, leisure and work. They must have acquired their condition before age 18. Cerebral Palsy C2 Upper Severe to moderate quadriplegia. Upper extremity events are Wheelchair performed by pushing the wheelchair with one or two arms and the wheelchair propulsion is restricted due to poor control. Upper extremity athletes have limited control of movements, but are able to produce some semblance of throwing motion. T/F 33 C3 Wheelchair Moderate quadriplegia. Fair functional strength and moderate problems in upper extremities and torso. -

Structure of the Full Kinetoplastids Mitoribosome and Insight on Its Large Subunit Maturation
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.02.073890; this version posted June 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Structure of the full kinetoplastids mitoribosome and insight on its large subunit maturation Heddy Soufari1* & Florent Waltz1*, Camila Parrot1+, Stéphanie Durrieu1+, Anthony Bochler1, Lauriane Kuhn2, Marie Sissler1, Yaser Hashem1‡ 1 Institut Européen de Chimie et Biologie, U1212 Inserm, UMR5320 CNRS, Université de Bordeaux, 2 rue R. Escarpit, F-33600 Pessac, France 2 Plateforme protéomique Strasbourg Esplanade FRC1589 du CNRS, Université de Strasbourg, Strasbourg, France *contributed equally, co-first authors +contributed equally, co-second authors ‡corresponding author Abstract: Kinetoplastids are unicellular eukaryotic parasites responsible for human pathologies such as Chagas disease, sleeping sickness or Leishmaniasis1. They possess a single large mitochondrion, essential for the parasite survival2. In kinetoplastids mitochondrion, most of the molecular machineries and gene expression processes have significantly diverged and specialized, with an extreme example being their mitochondrial ribosomes3. These large complexes are in charge of translating the few essential mRNAs encoded by mitochondrial genomes4,5. Structural studies performed in Trypanosoma brucei already highlighted the numerous peculiarities of these mitoribosomes and the maturation of their small subunit3,6. However, several important aspects mainly related to the large subunit remain elusive, such as the structure and maturation of its ribosomal RNA3. Here, we present a cryo-electron microscopy study of the protozoans Leishmania tarentolae and Trypanosoma cruzi mitoribosomes. -

S41467-017-00439-1.Pdf
ARTICLE DOI: 10.1038/s41467-017-00439-1 OPEN Biosynthesis of the nosiheptide indole side ring centers on a cryptic carrier protein NosJ Wei Ding1,2,3,4, Wenjuan Ji2, Yujie Wu2,3, Runze Wu2, Wan-Qiu Liu2, Tianlu Mo2, Junfeng Zhao2, Xiaoyan Ma2,3, Wei Zhang4, Ping Xu5, Zixin Deng6, Boping Tang1,YiYu6 & Qi Zhang 2 Nosiheptide is a prototypal thiopeptide antibiotic, containing an indole side ring in addition to its thiopeptide-characteristic macrocylic scaffold. This indole ring is derived from 3-methyl-2- indolic acid (MIA), a product of the radical S-adenosylmethionine enzyme NosL, but how MIA is incorporated into nosiheptide biosynthesis remains to be investigated. Here we report functional dissection of a series of enzymes involved in nosiheptide biosynthesis. We show NosI activates MIA and transfers it to the phosphopantetheinyl arm of a carrier protein NosJ. NosN then acts on the NosJ-bound MIA and installs a methyl group on the indole C4, and the resulting dimethylindolyl moiety is released from NosJ by a hydrolase-like enzyme NosK. Surface plasmon resonance analysis show that the molecular complex of NosJ with NosN is much more stable than those with other enzymes, revealing an elegant biosynthetic strategy in which the reaction flux is controlled by protein–protein interactions with different binding affinities. 1 Jiangsu Key Laboratory for Bioresources of Saline Soils, Jiangsu Synthetic Innovation Center for Coastal Bioagriculture, Yancheng Teachers University, Yancheng 224002, China. 2 Department of Chemistry, Fudan University, Shanghai 200433, China. 3 Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou 730000, China. -

Structure of the Full Kinetoplastids Mitoribosome and Insight on Its Large Subunit Maturation
Structure of the full kinetoplastids mitoribosome and insight on its large subunit maturation Heddy Soufari, Florent Waltz, Camila Parrot, Stéphanie Durrieu, Anthony Bochler, Lauriane Kuhn, Marie Sissler, Yaser Hashem To cite this version: Heddy Soufari, Florent Waltz, Camila Parrot, Stéphanie Durrieu, Anthony Bochler, et al.. Structure of the full kinetoplastids mitoribosome and insight on its large subunit maturation. 2020. hal-03000762 HAL Id: hal-03000762 https://hal.archives-ouvertes.fr/hal-03000762 Preprint submitted on 12 Nov 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. bioRxiv preprint doi: https://doi.org/10.1101/2020.05.02.073890; this version posted June 30, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Structure of the full kinetoplastids mitoribosome and insight on its large subunit maturation Heddy Soufari1* & Florent Waltz1*, Camila Parrot1+, Stéphanie Durrieu1+, Anthony Bochler1, Lauriane Kuhn2, Marie Sissler1, Yaser Hashem1‡ 1 Institut Européen de Chimie et Biologie, U1212 Inserm, UMR5320 CNRS, Université de Bordeaux, 2 rue R. -

RULES 2004 Final Amended
INTERNATIONAL PARALYMPIC COMMITTEE ATHLETICS SECTION COMBINED RULES 2004 PREAMBLE For competition at Paralympics and I.P.C. World Championships, this book shall be used, along with the current I.A.A.F. Handbook. It contains all the rules, which govern an I.P.C. Athletics competition, written in a way, which is compatible with the rules of the governing body for athletics, the International Association of Athletics Federations (I.A.A.F.). In this way, officials, coaches and athletes may find rules to cover any event in a single document, rather than having to refer to separate books for each group. The rules must be read in conjunction with the I.A.A.F. rules, contained in the Handbook of that Association. For the period including the 2004 Athens Paralympics, the version of the I.A.A.F. Handbook to which this book refers is the 2004-2005 edition. The rules concerned are the Technical Rules of Competition as described herein, and additional rules as shown. The reference to the I.A.A.F. Handbook does not confer any responsibility onto the I.A.A.F. for the I.P.C. Rules. Each of the I.O.S.D.’s has its own cycle of rule changes, and this volume does not intend to change any rules so decided. It is quite simply a means of simplifying the task of reading the rules for those concerned. NOTES This Rule Book will remain in force until the publication of the next I.A.A.F. Handbook, at which time a new edition will be published to take account of any new I.A.A.F. -

Information to Users
INFORMATION TO USERS While the most advanced technology has heen used to photograph and reproduce this manuscript, the qualify of the reproduction is heavily dependent upon the qualify of the material submitted. For example: • Manuscript pages may have indistinct print. In such cases, the best available copy has been filmed. • Manuscripts may not always be complete. In such cases, a note will indicate that it is not possible to obtain missing pages. • Copyrighted material may have been removed from the manuscript. In such cases, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, and charts) are photographed by sectioning the origined, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. Each oversize page is also filmed as one exposure and is available, for an additional charge, as a standard 35mm slide or as a 17”x 23” black and white photographic print. Most photographs reproduce acceptably on positive microfilm or microfiche but lack the clarify on xerographic copies made from the microfilm. For an additional charge, 35mm slides of 6”x 9” black and white photographic prints are available for any photographs or illustrations that cannot he reproduced satisfactorily by xerography. 8703560 Houck, David Renwick STUDIES ON THE BIOSYNTHESIS OF THE MODIFIED-PEPTIDE ANTIBIOTIC, NOSIHEPTIDE The Ohio State University Ph.D. 1986 University Microfilms I nternetionsi!300 N. Zeeb R oad, Ann Arbor, Ml 48106 STUDIES ON THE BIOSYNTHESIS OF THE MODIFIED-PEPTIDE ANTIBIOTIC, NOSIHEPTIDE DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By David Renwick Houck, B.S., M.S. -

Tatiara PWA Numerical Groundwater Flow Model and Projected Scenarios: Volume 1
Tatiara PWA numerical groundwater flow model and projected scenarios: Volume 1 Chris Li and Roger Cranswick Department of Environment, Water and Natural Resources September, 2017 DEWNR Technical report 2017/17 Department for Environment and Water GPO Box 1047, Adelaide SA 5001 Telephone National (08) 8463 6946 International +61 8 8463 6946 Fax National (08) 8463 6999 International +61 8 8463 6999 Website www.environment.sa.gov.au Disclaimer The Department for Environment and Water and its employees do not warrant or make any representation regarding the use, or results of the use, of the information contained herein as regards to its correctness, accuracy, reliability, currency or otherwise. The Department for Environment and Water and its employees expressly disclaims all liability or responsibility to any person using the information or advice. Information contained in this document is correct at the time of writing. Licensed under Creative Commons Attribution 4.0 Australia License © Crown in right of the State of South Australia through the Department for Environment and Water 2018 ISBN 978-1-925668-33-9 Preferred way to cite this publication Li C and RH Cranswick, 2017. Tatiara PWA numerical groundwater flow model and scenario projections: Volume 1. DEWNR Technical report 2017/17, Government of South Australia, Department of Environment, Water and Natural Resources, Adelaide. Download this document at https:// www.waterconnect.sa.gov.au DEWNR Technical report 2017/17 i Foreword The Department for Environment and Water (DEW) is responsible for the management of the State’s natural resources, ranging from policy leadership to on-ground delivery in consultation with government, industry and communities. -

The Funcsearch Search Engine
Improving search engine technology (Master thesis) Paul de Vrieze s920851 7-Mar-2002 Abstract While searching the internet for textual pages works rather well, searching for interactive pages can sometimes be still problematic. This thesis presents a solution to this problem. The solution presented uses a classification system that classifies pages into different classes to extend search engine functionality. Possible classifications are: textual web pages, link pages, and interactive pages. The classification system specifically improves searching interactive pages. To explain that, first the difference between textual web-pages and interactive web-pages needs to be explained. In textual web-pages the more important parts of the page are formed by words. In interactive pages, tags basically determine the structure of the page. Traditional search engines are based upon information retrieval. Information retrieval is mainly designed for textual contents (and not html specific). As in interactive pages tags play a big role, the traditional approach works less than perfect. Most literature that looks at search engines tries to improve searching textual pages. Others do try to improve searching other kinds of pages. They do though look for the solution at metadata. They want to search metadata for wanted pages. While this idea works very well there is one problem. There is as yet almost no metadata publicly available about web-pages. This metadata should be created by the authors of the web-pages. As it is possible that authors of large sites will provide metadata, small web-sites probably won't in the forseeable future. To improve searching for interactive pages a classification system can be used. -

Multidrug Recognition PNAS PLUS in a Minimal Bacterial Multidrug Resistance System
Structural basis and dynamics of multidrug recognition PNAS PLUS in a minimal bacterial multidrug resistance system Judith Habazettla, Martin Allana,1, Pernille Rose Jensena,2, Hans-Jürgen Sassa, Charles J. Thompsonb, and Stephan Grzesieka,3 aFocal Area Structural Biology and Biophysics, Biozentrum, University of Basel, CH-4056 Basel, Switzerland; and bDepartment of Microbiology and Immunology, Life Sciences Center, University of British Columbia, Vancouver, British Columbia V6T 1Z3, Canada Edited by Adriaan Bax, National Institutes of Health, Bethesda, MD, and approved November 11, 2014 (received for review June 27, 2014) TipA is a transcriptional regulator found in diverse bacteria. It con- induces folding of a larger, intrinsically unstructured part of stitutes a minimal autoregulated multidrug resistance system the protein (24). By this mechanism, TipA recognizes a variety of against numerous thiopeptide antibiotics. Here we report the thiopeptide antibiotics such as thiostrepton, nosiheptide, or structures of its drug-binding domain TipAS in complexes with promothiocin A, and confers resistance via up-regulation of the promothiocin A and nosiheptide, and a model of the thiostrepton tipA gene and possibly other MDR systems (23–25). The thio- complex. Drug binding induces a large transition from a partially peptide antibiotics induce expression of the tipA gene as two unfolded to a globin-like structure. The structures rationalize the alternate in-frame translation products: a long protein, TipAL mechanism of promiscuous, yet specific, drug recognition: (i)a (253 aa), and a short protein, TipAS (144 aa), which also con- four-ring motif present in all known TipA-inducing antibiotics is stitutes the C-terminal part of TipAL (24, 26). -

Syntheses of Fused Pyrroloheterocycles, Isatins, Approach Towards the Indole Fragment of Nosiheptide and a Base-Mediated Formation of 3-Hydroxycarbazoles
Graduate Theses, Dissertations, and Problem Reports 2009 Syntheses of fused pyrroloheterocycles, isatins, approach towards the indole fragment of nosiheptide and a base-mediated formation of 3-hydroxycarbazoles Sobha Priyadarshini Gorugantula West Virginia University Follow this and additional works at: https://researchrepository.wvu.edu/etd Recommended Citation Gorugantula, Sobha Priyadarshini, "Syntheses of fused pyrroloheterocycles, isatins, approach towards the indole fragment of nosiheptide and a base-mediated formation of 3-hydroxycarbazoles" (2009). Graduate Theses, Dissertations, and Problem Reports. 2851. https://researchrepository.wvu.edu/etd/2851 This Dissertation is protected by copyright and/or related rights. It has been brought to you by the The Research Repository @ WVU with permission from the rights-holder(s). You are free to use this Dissertation in any way that is permitted by the copyright and related rights legislation that applies to your use. For other uses you must obtain permission from the rights-holder(s) directly, unless additional rights are indicated by a Creative Commons license in the record and/ or on the work itself. This Dissertation has been accepted for inclusion in WVU Graduate Theses, Dissertations, and Problem Reports collection by an authorized administrator of The Research Repository @ WVU. For more information, please contact [email protected]. Syntheses of Fused Pyrroloheterocycles, Isatins, Approach Towards the Indole Fragment of Nosiheptide and a Base-Mediated Formation of 3-Hydroxycarbazoles Sobha Priyadarshini Gorugantula Dissertation submitted to the Eberly College of Arts and Sciences at West Virginia University in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Chemistry Björn C. G. Söderberg, Ph.D., Chair Patrick S. -

Deportistas Sin Adjetivos
Part 1-0 AUTORES-INDICE-INTRO:Part 1-0 AUTORES-INDICE-INTRO.qxd 12/12/2011 22:30 Página 5 Part 1-0 AUTORES-INDICE-INTRO:Part 1-0 AUTORES-INDICE-INTRO.qxd 12/12/2011 19:59 Página 6 Edita Consejo Superior de Deportes, Real Patronato sobre Discapacidad. Ministerio de Sanidad, Política Social e Igualdad Comité Paralímpico Español Consejo de Redacción Joan Palau Francàs. Presidente de la FEDDF Josep Oriol Martínez i Ferrer. Adjunto al Presidente de la FEDDF Miguel Ángel García Alfaro. Gerente y Director Técnico de la FEDDF Merche Ríos Hernández. Profesora de la Universitat de Barcelona Dirección editorial Javier Fernández Diseño y maquetación Ana F. Pérez Producción G·2 · [email protected] Impresión Croma graf Press c. o. NIPO (CSD): 008-11-021-13 NIPO (RPD): 864-11-015-6 Depósito Legal: M-46953 -2011 Todos los derechos reservados, no pudiéndose utilizar mecánica ni electrónicamente parte o la totalidad de la obra sin permiso de sus autores o del editor. Será requerido quien infrinja las normas legales establecidas. Impreso en España · Printed in Spain Part 1-0 AUTORES-INDICE-INTRO:Part 1-0 AUTORES-INDICE-INTRO.qxd 02/12/2011 19:39 Página 7 Esta Obra, que ha tenido un largo periodo de confección, ha sido posible gracias al aporte e impulso de una figura seguramente irrepetible en el mundo del deporte español, que por desgracia nos dejó el 26 de abril de 2011, Joan Palau, a quien todos los que hemos colaborado en la misma queremos brindar su puesta de largo. Esto es por y para ti. -

Uk Athletics Rules for Competition
UKA RULES FOR COMPETITION Effective from 1st April 2014 UK Athletics Athletics House Alexander Stadium Walsall Road Perry Barr Birmingham B42 2BE Lynn Davies, C.B.E. - President Edmond Warner O.B.E. - Chair Niels de Vos - Chief Executive Tel 0121 713 8400 Fax 0121 713 8452 W: www.britishathletics.org.uk E: [email protected] UK Athletics Ltd. ISBN: 978-0-9547401-3-9 Typeset by Data Standards Ltd., Frome Printed and bound by Polestar Wheatons Ltd., Exeter OFFICERS OF UKA GROUPS OFFICIALS GROUP Coordinator: Andrew Clatworthy, 26 Columba Drive, Leighton Buzzard, Beds. LU7 3YN. Tel/Fax: 01525 371186; E: [email protected] NATIONAL ASSOCIATIONS ENGLAND ATHLETICS Office: Alexander Stadium, Walsall Road, Perry Barr, Birmingham. B42 2BE.Tel: 0121 347 6543. Fax: 0121 347 6544. www.englandathletics.org Tri-Regional Officials’ Secretaries: South: Mrs Wendy Haxell, 5 Nursery Gardens, Horndean, Waterlooville. PO8 9LE. Tel: 02392 613717. E: [email protected]. Midlands & South West: Barry Parker, 26 Field Drive, Alvaston, Derby. DE24 0HF. Tel: 01332 753933; E: [email protected]. North: Paul Yates, 29 Chadwick Road, Eccles, Lancashire. M30 0WU. Tel: 07905 340077. E; [email protected]. ATHLETICS NORTHERN IRELAND Office: Athletics House, Old Coach Road, Belfast. BT79 5PR. Tel: 028 9060 2707; Fax: 028 9030 9939. www.niathletics.org. Officials’ Secretary: Bob Brodie, 3 Millar’s Ford, Lisburn, Northern Ireland. BT28 1JY. Tel: 028 9267 0311; Mob: 07939 670371; E: [email protected]. SCOTTISH ATHLETICS LTD Office: Caledonia House, South Gyle, Edinburgh. EH12 9DQ. Tel: 0131 539 7320. www.scottishathletics.org.uk Officials’ Convenor: Vic Hockley, 41 Murieston Park, Murieston South, Livingston, West Lothian.