Virus-Host Interactomics: New Insights and Opportunities for Antiviral Drug Discovery
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Treatment of Herpes Simplex Virus Epithelial Keratitis
THE TREATMENT OF HERPES SIMPLEX VIRUS EPITHELIAL KERATITIS BY Kirk R. Wilhelmus, MD, MPH ABSTRACT Purpose: Epithelial keratitis is the most common presentation of ocular infection by herpes simplex virus (HSV). Quantitative assessment of available therapy is needed to guide evidence-based ophthalmology. This study aimed to compare the efficacy of various treatments for dendritic or geographic HSV epithelial keratitis and to evaluate the role of various clinical characteristics on epithelial healing. Methods: Following a systematic review of the literature, information from clinical trials of HSV dendritic or geographic epithelial keratitis was extracted, and the methodological quality of each study was scored. Methods of epithelial cau- terization and curettage were grouped as relatively equivalent physicochemical therapy, and solution and ointment for- mulations of a given topical antiviral agent were combined. The proportion healed with 1 week of therapy, a scheduled follow-up day that approximated the average time of resolution with antiviral therapy, was selected as the primary out- come based on a masked evaluation of maximum treatment differences in published healing curves. The proportion healed at 14 days was recorded as supplemental information. Fixed-effects and random-effects meta-analysis models were used to obtain summary estimates by pooling results from comparative treatment trials. Hypotheses about which prognostic factors might affect epithelial healing during antiviral therapy were developed by multivariate analysis of the Herpetic Eye Disease Study dataset. Results: After excluding 48 duplicate reports, 14 nonrandomized studies, 15 studies with outdated or similar treatments, and 29 trials lacking sufficient data on healing or accessibility, 76 primary reports were identified. -
(12) United States Patent (10) Patent No.: US 8,993,581 B2 Perrine Et Al
US00899.3581B2 (12) United States Patent (10) Patent No.: US 8,993,581 B2 Perrine et al. (45) Date of Patent: Mar. 31, 2015 (54) METHODS FOR TREATINGVIRAL (58) Field of Classification Search DSORDERS CPC ... A61K 31/00; A61K 31/166; A61K 31/185: A61K 31/233; A61K 31/522: A61K 38/12: (71) Applicant: Trustees of Boston University, Boston, A61K 38/15: A61K 45/06 MA (US) USPC ........... 514/263.38, 21.1, 557, 565, 575, 617; 424/2011 (72) Inventors: Susan Perrine, Weston, MA (US); Douglas Faller, Weston, MA (US) See application file for complete search history. (73) Assignee: Trustees of Boston University, Boston, (56) References Cited MA (US) U.S. PATENT DOCUMENTS (*) Notice: Subject to any disclaimer, the term of this 3,471,513 A 10, 1969 Chinn et al. patent is extended or adjusted under 35 3,904,612 A 9/1975 Nagasawa et al. U.S.C. 154(b) by 0 days. (Continued) (21) Appl. No.: 13/915,092 FOREIGN PATENT DOCUMENTS (22) Filed: Jun. 11, 2013 CA 1209037 A 8, 1986 CA 2303268 A1 4f1995 (65) Prior Publication Data (Continued) US 2014/OO45774 A1 Feb. 13, 2014 OTHER PUBLICATIONS Related U.S. Application Data (63) Continuation of application No. 12/890,042, filed on PCT/US 10/59584 Search Report and Written Opinion mailed Feb. Sep. 24, 2010, now abandoned. 11, 2011. (Continued) (60) Provisional application No. 61/245,529, filed on Sep. 24, 2009, provisional application No. 61/295,663, filed on Jan. 15, 2010. Primary Examiner — Savitha Rao (74) Attorney, Agent, or Firm — Nixon Peabody LLP (51) Int. -
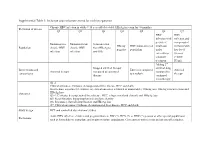
Inclusion and Exclusion Criteria for Each Key Question
Supplemental Table 1: Inclusion and exclusion criteria for each key question Chronic HBV infection in adults ≥ 18 year old (detectable HBsAg in serum for >6 months) Definition of disease Q1 Q2 Q3 Q4 Q5 Q6 Q7 HBV HBV infection with infection and persistent compensated Immunoactive Immunotolerant Seroconverted HBeAg HBV mono-infected viral load cirrhosis with Population chronic HBV chronic HBV from HBeAg to negative population under low level infection infection anti-HBe entecavir or viremia tenofovir (<2000 treatment IU/ml) Adding 2nd Stopped antiviral therapy antiviral drug Interventions and Entecavir compared Antiviral Antiviral therapy compared to continued compared to comparisons to tenofovir therapy therapy continued monotherapy Q1-2: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Intermediate outcomes (if evidence on clinical outcomes is limited or unavailable): HBsAg loss, HBeAg seroconversion and Outcomes HBeAg loss Q3-4: Cirrhosis, decompensated liver disease, HCC, relapse (viral and clinical) and HBsAg loss Q5: Renal function, hypophosphatemia and bone density Q6: Resistance, flare/decompensation and HBeAg loss Q7: Clinical outcomes: Cirrhosis, decompensated liver disease, HCC and death Study design RCT and controlled observational studies Acute HBV infection, children and pregnant women, HIV (+), HCV (+) or HDV (+) persons or other special populations Exclusions such as hemodialysis, transplant, and treatment failure populations. Co treatment with steroids and uncontrolled studies. Supplemental Table 2: Detailed Search Strategy: Ovid Database(s): Embase 1988 to 2014 Week 37, Ovid MEDLINE(R) In-Process & Other Non- Indexed Citations and Ovid MEDLINE(R) 1946 to Present, EBM Reviews - Cochrane Central Register of Controlled Trials August 2014, EBM Reviews - Cochrane Database of Systematic Reviews 2005 to July 2014 Search Strategy: # Searches Results 1 exp Hepatitis B/dt 26410 ("hepatitis B" or "serum hepatitis" or "hippie hepatitis" or "injection hepatitis" or 2 178548 "hepatitis type B").mp. -

Review of Sezary Syndrome
REVIEWS Sezary syndrome: Immunopathogenesis, literature review of therapeutic options, and recommendations for therapy by the United States Cutaneous Lymphoma Consortium (USCLC) EliseA.Olsen,MD,a Alain H. Rook, MD,b John Zic, MD,c Youn Kim, MD,d PierluigiPorcu,MD,e Christiane Querfeld, MD,f Gary Wood, MD,g Marie-France Demierre, MD,h Mark Pittelkow, MD,i Lynn D. Wilson, MD, MPH,j Lauren Pinter-Brown, MD,k Ranjana Advani, MD,d Sareeta Parker, MD,l Ellen J. Kim, MD,b Jacqueline M. Junkins-Hopkins, MD,m Francine Foss, MD,j Patrick Cacchio, BS,a and Madeleine Duvic, MDn Durham, North Carolina; Philadelphia, Pennsylvania; Nashville, Tennessee; Palo Alto and Los Angeles, California; Columbus, Ohio; Chicago, Illinois;Madison,Wisconsin;Boston,Massachusetts; Rochester, Minnesota; New Haven, Connecticut; Atlanta, Georgia; Baltimore, Maryland; and Houston, Texas Sezary syndrome (SS) has a poor prognosis and few guidelines for optimizing therapy. The US Cutaneous Lymphoma Consortium, to improve clinical care of patients with SS and encourage controlled clinical trials of promising treatments, undertook a review of the published literature on therapeutic options for SS. An overview of the immunopathogenesis and standardized review of potential current treatment options for SS including metabolism, mechanism of action, overall efficacy in mycosis fungoides and SS, and common or concerning adverse effects is first discussed. The specific efficacy of each treatment for SS, both as monotherapy and combination therapy, is then reported using standardized criteria for both SS and response to therapy with the type of study defined by a modification of the US Preventive Services guidelines for evidence-based medicine. -

Emerging Role of Mucosal Vaccine in Preventing Infection with Avian Influenza a Viruses
viruses Review Emerging Role of Mucosal Vaccine in Preventing Infection with Avian Influenza A Viruses Tong Wang 1, Fanhua Wei 2 and Jinhua Liu 1,* 1 Key Laboratory of Animal Epidemiology and Zoonosis, Ministry of Agriculture, College of Veterinary Medicine, China Agricultural University, Beijing 100193, China; [email protected] 2 College of Agriculture, Ningxia University, Yinchuan 750021, China; [email protected] * Correspondence: [email protected] Received: 22 July 2020; Accepted: 5 August 2020; Published: 7 August 2020 Abstract: Avian influenza A viruses (AIVs), as a zoonotic agent, dramatically impacts public health and the poultry industry. Although low pathogenic avian influenza virus (LPAIV) incidence and mortality are relatively low, the infected hosts can act as a virus carrier and provide a resource pool for reassortant influenza viruses. At present, vaccination is the most effective way to eradicate AIVs from commercial poultry. The inactivated vaccines can only stimulate humoral immunity, rather than cellular and mucosal immune responses, while failing to effectively inhibit the replication and spread of AIVs in the flock. In recent years, significant progresses have been made in the understanding of the mechanisms underlying the vaccine antigen activities at the mucosal surfaces and the development of safe and efficacious mucosal vaccines that mimic the natural infection route and cut off the AIVs infection route. Here, we discussed the current status and advancement on mucosal immunity, the means of establishing mucosal immunity, and finally a perspective for design of AIVs mucosal vaccines. Hopefully, this review will help to not only understand and predict AIVs infection characteristics in birds but also extrapolate them for distinction or applicability in mammals, including humans. -

The Phytochemistry of Cherokee Aromatic Medicinal Plants
medicines Review The Phytochemistry of Cherokee Aromatic Medicinal Plants William N. Setzer 1,2 1 Department of Chemistry, University of Alabama in Huntsville, Huntsville, AL 35899, USA; [email protected]; Tel.: +1-256-824-6519 2 Aromatic Plant Research Center, 230 N 1200 E, Suite 102, Lehi, UT 84043, USA Received: 25 October 2018; Accepted: 8 November 2018; Published: 12 November 2018 Abstract: Background: Native Americans have had a rich ethnobotanical heritage for treating diseases, ailments, and injuries. Cherokee traditional medicine has provided numerous aromatic and medicinal plants that not only were used by the Cherokee people, but were also adopted for use by European settlers in North America. Methods: The aim of this review was to examine the Cherokee ethnobotanical literature and the published phytochemical investigations on Cherokee medicinal plants and to correlate phytochemical constituents with traditional uses and biological activities. Results: Several Cherokee medicinal plants are still in use today as herbal medicines, including, for example, yarrow (Achillea millefolium), black cohosh (Cimicifuga racemosa), American ginseng (Panax quinquefolius), and blue skullcap (Scutellaria lateriflora). This review presents a summary of the traditional uses, phytochemical constituents, and biological activities of Cherokee aromatic and medicinal plants. Conclusions: The list is not complete, however, as there is still much work needed in phytochemical investigation and pharmacological evaluation of many traditional herbal medicines. Keywords: Cherokee; Native American; traditional herbal medicine; chemical constituents; pharmacology 1. Introduction Natural products have been an important source of medicinal agents throughout history and modern medicine continues to rely on traditional knowledge for treatment of human maladies [1]. Traditional medicines such as Traditional Chinese Medicine [2], Ayurvedic [3], and medicinal plants from Latin America [4] have proven to be rich resources of biologically active compounds and potential new drugs. -

Herpes Simplex Virus
HSV Herpes simplex virus HSV (Herpes simplex virus) can be spread when an infected person is producing and shedding the virus. Herpes simplex can be spread through contact with saliva, such as sharing drinks. Symptoms of herpes simplex virus infection include watery blisters in the skin or mucous membranes of the mouth, lips or genitals. Lesions heal with ascab characteristic of herpetic disease. As neurotropic and neuroinvasive viruses, HSV-1 and -2 persist in the body by becoming latent and hiding from the immune system in the cell bodies of neurons. After the initial or primary infection, some infected people experience sporadic episodes of viral reactivation or outbreaks. www.MedChemExpress.com 1 HSV Inhibitors (Z)-Capsaicin 1-Docosanol (Zucapsaicin; Civamide; cis-Capsaicin) Cat. No.: HY-B1583 (Behenyl alcohol) Cat. No.: HY-B0222 (Z)-Capsaicin is the cis isomer of capsaicin, acts 1-Docosanol is a saturated fatty alcohol used as an orally active TRPV1 agonist, and is used in traditionally as an emollient, emulsifier, and the research of neuropathic pain. thickener in cosmetics, and nutritional supplement; inhibitor of lipid-enveloped viruses including herpes simplex. Purity: 99.96% Purity: ≥98.0% Clinical Data: Launched Clinical Data: Launched Size: 10 mM × 1 mL, 10 mg, 50 mg Size: 500 mg 2-Deoxy-D-glucose 20(R)-Ginsenoside Rh2 (2-DG; 2-Deoxy-D-arabino-hexose; D-Arabino-2-deoxyhexose) Cat. No.: HY-13966 Cat. No.: HY-N1401 2-Deoxy-D-glucose is a glucose analog that acts as 20(R)-Ginsenoside Rh2, a matrix a competitive inhibitor of glucose metabolism, metalloproteinase (MMP) inhibitor, acts as a inhibiting glycolysis via its actions on hexokinase. -

View/Download
Research Article Research Article Dermatology Research Therapy with Pegylated Interferon or Combined with Cryosurgery in Condyloma Acuminata. Phase III Clinical Trial Israel Alfonso Trujillo*, Tomás Tabilo Bocic, Ángela Rosa Gutiérrez Rojas, Hugo Nodarse Cuní, María Elena Flores Andrade, and María del Carmen Toledo García *Correspondence: Israel Alfonso-Trujillo, Calzada of Managua # 1133 e/Caimán and Quemados. Las Guásimas. Arroyo Naranjo, Havana, Cuba, E-mail: Surgical Clinical Hospital: "Hermanos Ameijeiras", Cuba. [email protected] Received: 04 January 2019; Accepted: 18 February 2019 Citation: Israel Alfonso Trujillo, Tomás Tabilo Bocic, Ángela Rosa Gutiérrez Rojas, et al. Therapy with Pegylated Interferon or Combined with Cryosurgery in Condyloma Acuminata. Phase III Clinical Trial. Dermatol Res. 2019; 1(1); 1-8. ABSTRACT Background: The continuous recurrence of condyloma acuminata makes the constant search for necessary therapeutic alternatives. Patients and method: To evaluate the therapeutic efficacy and safety of pegylated interferon, alone or adjuvant for cryosurgery, in the condyloma acuminata an open clinical trial was carried out on 30 patients of the "Hermanos Ameijeiras" hospital, who were randomized to receive for 6 weeks (group A) only fortnightly cryosurgery, (group B) subcutaneous pegylated interferon, once a week, associated with fortnightly cryosurgery application or (group C) only subcutaneous pegylated interferon, once a week. The main variable was the percentage of recurrence at one year of follow-up, evaluated quarterly. There was also a rigorous control of adverse events. Results: At the end of the treatment 8/10 (80%) patients from group A, 10/10 (100%) from group B and 9/10 (90%) from group C were left without injuries. -

Belayachi Et Al., Afr J Tradit Complement Altern Med. (2017) 14(2):356-373 Doi:10.21010/Ajtcam.V14i2.37
Belayachi et al., Afr J Tradit Complement Altern Med. (2017) 14(2):356-373 doi:10.21010/ajtcam.v14i2.37 INDUCTION OF CELL CYCLE ARREST AND APOPTOSIS BY ORMENIS ERIOLEPIS A MORROCAN ENDEMIC PLANT IN VARIOUS HUMAN CANCER CELL LINES Lamiae Belayachia,b*, Clara Aceves-Luquerob, Nawel Merghoubd, Silvia Fernández de Mattosb,c, Saaïd a b,c a Amzazi , Priam Villalonga ,Youssef Bakri a Biochemistry-Immunology Laboratory, Faculty of Sciences, Mohammed V-Agdal University, Rabat, Morocco, b Cancer Cell Biology Group, Institut Universitari d’Investigació en Ciències de la Salut (IUNICS), c Departament de Biologia Fonamental, Universitat de les Illes Balears, Illes Balears, Spain, Green Biotechnology Center. MAScIR (Moroccan Foundation for Advanced Science, Innovation & Research)- Rabat Design Center, Rabat - Morocco. *Corresponding author E-mail: [email protected] Abstract Background: Ormenis eriolepis Coss (Asteraceae) is an endemic Moroccan subspecies, traditionally named “Hellala” or “Fergoga”. It’s usually used for its hypoglycemic effect as well as for the treatment of stomacal pain. As far as we know, there is no scientific exploration of anti tumoral activity of Ormenis eriolepis extracts. Materials and Methods: In this regard, we performed a screening of organic extracts and fractions in a panel of both hematological and solid cancer cell lines, to evaluate the potential in vitro anti tumoral activity and to elucidate the respective mechanisms that may be responsible for growth arrest and cell death induction. The plant was extracted using organic solvents, and four different extracts were screened on Jurkat, Jeko-1, TK-6, LN229, SW620, U2OS, PC-3 and NIH3T3 cells. Results: Cell viability assays revealed that, the IC50 values were (11,63±5,37µg/ml) for Jurkat, (13,33±1,67µg/ml) for Jeko-1, (41,67±1,98µg/ml) for LN229 and (19,31±4,88µg/ml) for PC-3 cells upon treatment with Oe-DF and Oe-HE respectively. -

National OTC Medicines List
National OTC Medicines List ‐ DraŌ 01 DRAFT National OTC Medicines List Draft 01 Ministry of Public Health of Lebanon This list was prepared under the guidance of His Excellency Minister Waêl Abou Faour andDRAFT the supervision of the Director General Dr. Walid Ammar. Editors Rita KARAM, Pharm D. PhD. Myriam WATFA, Pharm D Ghassan HAMADEH, MD.CPE FOREWORD According to the French National Agency for Medicines and Health Products Safety (ANSM), Over-the-counter (OTC) drugs are medicines that are accessible to patients in pharmacies, based on criteria set to safeguard patients’ safety. Due to their therapeutic class, these medicines could be dispensed without physician’s intervention for diagnostic, treatment initiation or maintenance purposes. Moreover, their dosage, treatment period and Package Insert Leaflet should be suitable for OTC classification. The packaging size should be in accordance with the dosage and treatment period. According to ArticleDRAFT 43 of the Law No.367 issued in 1994 related to the pharmacy practice, and the amendment of Articles 46 and 47 by Law No.91 issued in 2010, pharmacists do not have the right to dispense any medicine that is not requested by a unified prescription, unless the medicine is mentioned in a list which is established by pharmacists and physicians’ syndicates. In this regard, the Ministry of Public Health (MoPH) developed the National OTC Medicines List, and presentedit in a scientific, objective, reliable, and accessible listing. The OTC List was developed by a team of pharmacists and physicians from the Ministry of Public Health (MoPH). In order to ensure a safe and effective self- medicationat the pharmacy level, several pharmaceutical categories (e.g. -

Development and Effects of Influenza Antiviral Drugs
molecules Review Development and Effects of Influenza Antiviral Drugs Hang Yin, Ning Jiang, Wenhao Shi, Xiaojuan Chi, Sairu Liu, Ji-Long Chen and Song Wang * Key Laboratory of Fujian-Taiwan Animal Pathogen Biology, College of Animal Sciences (College of Bee Science), Fujian Agriculture and Forestry University, Fuzhou 350002, China; [email protected] (H.Y.); [email protected] (N.J.); [email protected] (W.S.); [email protected] (X.C.); [email protected] (S.L.); [email protected] (J.-L.C.) * Correspondence: [email protected] Abstract: Influenza virus is a highly contagious zoonotic respiratory disease that causes seasonal out- breaks each year and unpredictable pandemics occasionally with high morbidity and mortality rates, posing a great threat to public health worldwide. Besides the limited effect of vaccines, the problem is exacerbated by the lack of drugs with strong antiviral activity against all flu strains. Currently, there are two classes of antiviral drugs available that are chemosynthetic and approved against influenza A virus for prophylactic and therapeutic treatment, but the appearance of drug-resistant virus strains is a serious issue that strikes at the core of influenza control. There is therefore an urgent need to develop new antiviral drugs. Many reports have shown that the development of novel bioactive plant extracts and microbial extracts has significant advantages in influenza treat- ment. This paper comprehensively reviews the development and effects of chemosynthetic drugs, plant extracts, and microbial extracts with influenza antiviral activity, hoping to provide some refer- ences for novel antiviral drug design and promising alternative candidates for further anti-influenza drug development. -

Malta Medicines List April 08
Defined Daily Doses Pharmacological Dispensing Active Ingredients Trade Name Dosage strength Dosage form ATC Code Comments (WHO) Classification Class Glucobay 50 50mg Alpha Glucosidase Inhibitor - Blood Acarbose Tablet 300mg A10BF01 PoM Glucose Lowering Glucobay 100 100mg Medicine Rantudil® Forte 60mg Capsule hard Anti-inflammatory and Acemetacine 0.12g anti rheumatic, non M01AB11 PoM steroidal Rantudil® Retard 90mg Slow release capsule Carbonic Anhydrase Inhibitor - Acetazolamide Diamox 250mg Tablet 750mg S01EC01 PoM Antiglaucoma Preparation Parasympatho- Powder and solvent for solution for mimetic - Acetylcholine Chloride Miovisin® 10mg/ml Refer to PIL S01EB09 PoM eye irrigation Antiglaucoma Preparation Acetylcysteine 200mg/ml Concentrate for solution for Acetylcysteine 200mg/ml Refer to PIL Antidote PoM Injection injection V03AB23 Zovirax™ Suspension 200mg/5ml Oral suspension Aciclovir Medovir 200 200mg Tablet Virucid 200 Zovirax® 200mg Dispersible film-coated tablets 4g Antiviral J05AB01 PoM Zovirax® 800mg Aciclovir Medovir 800 800mg Tablet Aciclovir Virucid 800 Virucid 400 400mg Tablet Aciclovir Merck 250mg Powder for solution for inj Immunovir® Zovirax® Cream PoM PoM Numark Cold Sore Cream 5% w/w (5g/100g)Cream Refer to PIL Antiviral D06BB03 Vitasorb Cold Sore OTC Cream Medovir PoM Neotigason® 10mg Acitretin Capsule 35mg Retinoid - Antipsoriatic D05BB02 PoM Neotigason® 25mg Acrivastine Benadryl® Allergy Relief 8mg Capsule 24mg Antihistamine R06AX18 OTC Carbomix 81.3%w/w Granules for oral suspension Antidiarrhoeal and Activated Charcoal