Genetic Study of Dravidian Castes of Tamil Nadu
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Top 300 Masters 2020
TOP 300 MASTERS 2020 2020 Top 300 MASTERS 1 About Broadcom MASTERS Broadcom MASTERS® (Math, Applied Science, Technology and Engineering for Rising Stars), a program of Society for Science & the Public, is the premier middle school science and engineering fair competition, inspiring the next generation of scientists, engineers and innovators who will solve the grand challenges of the 21st century and beyond. We believe middle school is a critical time when young people identify their personal passion, and if they discover an interest in STEM, they can be inspired to follow their passion by taking STEM courses in high school. Broadcom MASTERS is the only middle school STEM competition that leverages Society- affiliated science fairs as a critical component of the STEM talent pipeline. In 2020, all 6th, 7th, and 8th grade students around the country who were registered for their local or state Broadcom MASTERS affiliated fair were eligible to compete. After submitting the online application, the Top 300 MASTERS are selected by a panel of scientists, engineers, and educators from around the nation. The Top 300 MASTERS are honored for their work with a $125 cash prize, through the Society’s partnership with the U.S. Department of Defense as a member of the Defense STEM education Consortium (DSEC). Top 300 MASTERS also receive a prize package that includes an award ribbon, a Top 300 MASTERS certificate of accomplishment, a Broadcom MASTERS backpack, a Broadcom MASTERS decal, a one-year family digital subscription to Science News magazine, an Inventor's Notebook, courtesy of The Lemelson Foundation, a one-year subscription to Wolfram Mathematica software, courtesy of Wolfram Research, and a special prize from Jeff Glassman, CEO of Covington Capital Management. -

Community List
ANNEXURE - III LIST OF COMMUNITIES I. SCHEDULED TRIB ES II. SCHEDULED CASTES Code Code No. No. 1 Adiyan 2 Adi Dravida 2 Aranadan 3 Adi Karnataka 3 Eravallan 4 Ajila 4 Irular 6 Ayyanavar (in Kanyakumari District and 5 Kadar Shenkottah Taluk of Tirunelveli District) 6 Kammara (excluding Kanyakumari District and 7 Baira Shenkottah Taluk of Tirunelveli District) 8 Bakuda 7 Kanikaran, Kanikkar (in Kanyakumari District 9 Bandi and Shenkottah Taluk of Tirunelveli District) 10 Bellara 8 Kaniyan, Kanyan 11 Bharatar (in Kanyakumari District and Shenkottah 9 Kattunayakan Taluk of Tirunelveli District) 10 Kochu Velan 13 Chalavadi 11 Konda Kapus 14 Chamar, Muchi 12 Kondareddis 15 Chandala 13 Koraga 16 Cheruman 14 Kota (excluding Kanyakumari District and 17 Devendrakulathan Shenkottah Taluk of Tirunelveli District) 18 Dom, Dombara, Paidi, Pano 15 Kudiya, Melakudi 19 Domban 16 Kurichchan 20 Godagali 17 Kurumbas (in the Nilgiris District) 21 Godda 18 Kurumans 22 Gosangi 19 Maha Malasar 23 Holeya 20 Malai Arayan 24 Jaggali 21 Malai Pandaram 25 Jambuvulu 22 Malai Vedan 26 Kadaiyan 23 Malakkuravan 27 Kakkalan (in Kanyakumari District and Shenkottah 24 Malasar Taluk of Tirunelveli District) 25 Malayali (in Dharmapuri, North Arcot, 28 Kalladi Pudukkottai, Salem, South Arcot and 29 Kanakkan, Padanna (in the Nilgiris District) Tiruchirapalli Districts) 30 Karimpalan 26 Malayakandi 31 Kavara (in Kanyakumari District and Shenkottah 27 Mannan Taluk of Tirunelveli District) 28 Mudugar, Muduvan 32 Koliyan 29 Muthuvan 33 Koosa 30 Pallayan 34 Kootan, Koodan (in Kanyakumari District and 31 Palliyan Shenkottah Taluk of Tirunelveli District) 32 Palliyar 35 Kudumban 33 Paniyan 36 Kuravan, Sidhanar 34 Sholaga 39 Maila 35 Toda (excluding Kanyakumari District and 40 Mala Shenkottah Taluk of Tirunelveli District) 41 Mannan (in Kanyakumari District and Shenkottah 36 Uraly Taluk of Tirunelveli District) 42 Mavilan 43 Moger 44 Mundala 45 Nalakeyava Code III (A). -

Persistence of Caste in South India - an Analytical Study of the Hindu and Christian Nadars
Copyright by Hilda Raj 1959 , PERSISTENCE OF CASTE IN SOUTH INDIA - AN ANALYTICAL STUDY OF THE HINDU AND CHRISTIAN NADARS by Hilda Raj Submitted to the Faculty of the Graduate School of The American University in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Signatures of Committee: . Chairman: D a t e ; 7 % ^ / < f / 9 < r f W58 7 a \ The American University Washington, D. 0. ACKNOWLEDGEMENTS I am deeply thankful to the following members of my Dissertation Committee for their guidance and sug gestions generously given in the preparation of the Dissertation: Doctors Robert T. Bower, N. G. D. Joardar, Lawrence Krader, Harvey C. Moore, Austin Van der Slice (Chairman). I express my gratitude to my Guru in Sociology, the Chairman of the above Committee - Dr. Austin Van der Slice, who suggested ways for the collection of data, and methods for organizing and presenting the sub ject matter, and at every stage supervised the writing of my Dissertation. I am much indebted to the following: Dr. Horace Poleman, Chief of the Orientalia Di vision of the Library of Congress for providing facilities for study in the Annex of the Library, and to the Staff of the Library for their unfailing courtesy and readi ness to help; The Librarian, Central Secretariat-Library, New Delhi; the Librarian, Connemara Public Library, Madras; the Principal in charge of the Library of the Theological Seminary, Nazareth, for privileges to use their books; To the following for helping me to gather data, for distributing questionnaire forms, collecting them after completion and mailing them to my address in Washington: Lawrence Gnanamuthu (Bombay), Dinakar Gnanaolivu (Madras), S. -

Agastya International Foundation Synopsys
In Partnership with Student Projects Technical Record Released on the occasion of Science & Engineering Fair of Selected Projects at Shikshakara Sadana, K G Road Bangalore on th th th 26 , 27 & 28 February 2018 Organised by Agastya International Foundation In support with Synopsys Anveshana 2017-18- BANGALORE - Abstract Book Page 1 of 209 CONTENTS 1. FOREWORD 2. ABOUT AGASTYA INTERNATIONAL FOUNDATION 3. ABOUT SYNOPSYS 4. ABOUT ANVESHANA 5. PROJECT SCREENING COMMITTEE 6. COPY OF INVITATION 7. PROGRAM CHART 8. LIST OF PROJECTS EXHIBITED IN THE FAIR 9. PROJECT DESCRIPTION Anveshana 2017-18- BANGALORE - Abstract Book Page 2 of 209 FOREWORD In a world where recent events suggest that we may be entering a period of greater uncertainties, it is disturbing that India's educational system is not (in general) internationally competitive. In an age where the state of the economy is driven more and more by knowledge and skill, it is clear that the future of our country will depend crucially on education at all levels – from elementary schools to research universities. It is equally clear that the question is not one of talent or innate abilities of our country men, as more and more Indians begin to win top jobs in US business and industry, government and academia. Indian talent is almost universally acknowledged, as demonstrated by the multiplying number of R&D centres being set up in India by an increasing number of multinational companies. So what is the real problem? There are many problems ranging from poor talent management to an inadequate teaching system in most schools and colleges where there is little effort to make contact with the real world in general rather than only prescribed text books. -

Nadarin Pdf, Epub, Ebook
NADARIN PDF, EPUB, EBOOK Leo Lionni,Teresa Mlawer | 24 pages | 30 Sep 2005 | Lectorum Publications | 9781930332805 | English, Spanish | New York, NY, United States Nadarin PDF Book Swimmy , A. Now customize the name of a clipboard to store your clips. They were also very caste conscious. The support included urging their own community members to allow use of their schools, tanks, temples and wells by other communities. Any Condition Any Condition. Impersonate a cat, then when questioned again say you're a stray. She wants you to retrieve an amulet from Arkion. Raid the trapped drawers and steal the Rogue Stone. About this product. Namespaces Article Talk. In the early nineteenth century, the Nadars were a community mostly engaged in the palmyra industry, including the production of toddy. Kamaraj , whose opinions had originally been disliked by his own community. Hardgrave conjectures that the Nadars of Southern Travancore migrated there from Tirunelveli in the 16th century after the invasion of Tirunelveli by the Raja of Travancore. These include procedures relating to birth, adulthood, marriage and death. Password forgot password? Cookie Settings explains the different cookies we use. Forrester University of South Carolina press. In situations where the matter went to court, the Sangam would not provide financial support for the Nadar claimant to contest the case, but would rather see that the claim is properly heard. No notes for slide. Staycation by M. Please visit our current hours of operation for our outlets and amenities and a complete list of our enhanced hygiene safety measures. Additional information about this order and a list of high risk states may be found by clicking here. -

Linguistic History and Language Diversity in India: Views and Counterviews
J Biosci (2019) 44:62 Indian Academy of Sciences DOI: 10.1007/s12038-019-9879-1 (0123456789().,-volV)(0123456789().,-volV) Linguistic history and language diversity in India: Views and counterviews SONAL KULKARNI-JOSHI Deccan College, Pune, India (Email, [email protected]) This paper addresses the theme of the seminar from the perspective of historical linguistics. It introduces the construct of ‘language family’ and then proceeds to a discussion of contact and the dynamics of linguistic exchange among the main language families of India over several millennia. Some prevalent hypotheses to explain the creation of India as a linguistic area are presented. The ‘substratum view’ is critically assessed. Evidence from historical linguistics in support of two dominant hypotheses –‘the Aryan migration view’ and ‘the out-of-India hypothesis’–is presented and briefly assessed. In conclusion, it is observed that the current understanding in historical linguistics favours the Aryan migration view though the ‘substratum view’ is questionable. Keywords. Aryan migration; historical linguistics; language family; Out-of-India hypothesis; substratum 1. Introduction the basis of social, political and cultural criteria more than linguistic criteria. The aim of this paper is to lend a linguistic perspective on This vast number of languages is classified into four (or the issue of human diversity and ancestry in India to the non- six) language families or genealogical types: Austro-Asiatic linguists at this seminar. The paper is an overview of the (Munda), Dravidian, Indo-Aryan (IA) and Tibeto-Burman; major views and evidences gleaned from the available more recently, two other language families have been literature. -

Police Matters: the Everyday State and Caste Politics in South India, 1900�1975 � by Radha Kumar
PolICe atter P olice M a tte rs T he v eryday tate and aste Politics in South India, 1900–1975 • R a dha Kumar Cornell unIerIt Pre IthaCa an lonon Copyright 2021 by Cornell University The text of this book is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License: https:creativecommons.orglicensesby-nc-nd4.0. To use this book, or parts of this book, in any way not covered by the license, please contact Cornell University Press, Sage House, 512 East State Street, Ithaca, New ork 14850. Visit our website at cornellpress.cornell.edu. First published 2021 by Cornell University Press Library of Congress Cataloging-in-Publication Data Names: Kumar, Radha, 1981 author. Title: Police matters: the everyday state and caste politics in south India, 19001975 by Radha Kumar. Description: Ithaca New ork: Cornell University Press, 2021 Includes bibliographical references and index. Identifiers: LCCN 2021005664 (print) LCCN 2021005665 (ebook) ISBN 9781501761065 (paperback) ISBN 9781501760860 (pdf) ISBN 9781501760877 (epub) Subjects: LCSH: Police—India—Tamil Nadu—History—20th century. Law enforcement—India—Tamil Nadu—History—20th century. Caste— Political aspects—India—Tamil Nadu—History. Police-community relations—India—Tamil Nadu—History—20th century. Caste-based discrimination—India—Tamil Nadu—History—20th century. Classification: LCC HV8249.T3 K86 2021 (print) LCC HV8249.T3 (ebook) DDC 363.20954820904—dc23 LC record available at https:lccn.loc.gov2021005664 LC ebook record available at https:lccn.loc.gov2021005665 Cover image: The Car en Route, Srivilliputtur, c. 1935. The British Library Board, Carleston Collection: Album of Snapshot Views in South India, Photo 6281 (40). -

THE TEXTS, POLITICAL HISTORY and ADMINISTRATION Till C. 200 BC
History of Ancient India Volume III THE TEXTS, POLITICAL HISTORY AND ADMINISTRATION Till c. 200 BC Editors Dilip K. Chakrabarti and Makkhan Lal Vivekananda International Foundation New Delhi Aryan Books International New Delhi Cataloging in Publication Data—DK [Courtesy: D.K. Agencies (P) Ltd. <[email protected]>] History of ancient India / editors, Dilip K. Chakrabarti and Makkhan Lal. v. 3 cm. Contributed articles. Includes index. Contents: v. 3. The texts, political history and administration, till c. 200 BC. ISBN 9788173054822 1. India—History. 2. India—Politics and government. I. Chakrabarti, Dilip K., 1941- II. Makkhan Lal, 1954- III. Vivekananda International Foundation. DDC 954 23 ISBN: 978-81-7305-482-2 © Vivekananda International Foundation All rights reserved. No part of this book may be reproduced, utilised in any form or by any means, electronic and mechanical, including photocopying, recording or by any information storage and retrieval system without prior permission of the authors and the publishers. Responsibility for statements made and visuals provided in the various papers rest solely with the contributors. The views expressed by individual authors are not necessarily those of the editors or of publishers. First Published in 2014 by Vivekananda International Foundation 3, San Martin Marg, Chanakyapuri, New Delhi - 110 021 Tel.: 24121764, 24106698; Fax: 91-11-43115450 E-mail: [email protected] www.vifindia.org in association with Aryan Books International Pooja Apartments, 4B, Ansari Road, New Delhi - 110 002 Tel.: 23287589, 23255799; Fax: 91-11-23270385 E-mail: [email protected] www.aryanbooks.co.in Designed and Printed in India at ABI Prints & Publishing Co., New Delhi. -

Dictionary of Martyrs: India's Freedom Struggle
DICTIONARY OF MARTYRS INDIA’S FREEDOM STRUGGLE (1857-1947) Vol. 5 Andhra Pradesh, Telangana, Karnataka, Tamil Nadu & Kerala ii Dictionary of Martyrs: India’s Freedom Struggle (1857-1947) Vol. 5 DICTIONARY OF MARTYRSMARTYRS INDIA’S FREEDOM STRUGGLE (1857-1947) Vol. 5 Andhra Pradesh, Telangana, Karnataka, Tamil Nadu & Kerala General Editor Arvind P. Jamkhedkar Chairman, ICHR Executive Editor Rajaneesh Kumar Shukla Member Secretary, ICHR Research Consultant Amit Kumar Gupta Research and Editorial Team Ashfaque Ali Md. Naushad Ali Md. Shakeeb Athar Muhammad Niyas A. Published by MINISTRY OF CULTURE, GOVERNMENT OF IDNIA AND INDIAN COUNCIL OF HISTORICAL RESEARCH iv Dictionary of Martyrs: India’s Freedom Struggle (1857-1947) Vol. 5 MINISTRY OF CULTURE, GOVERNMENT OF INDIA and INDIAN COUNCIL OF HISTORICAL RESEARCH First Edition 2018 Published by MINISTRY OF CULTURE Government of India and INDIAN COUNCIL OF HISTORICAL RESEARCH 35, Ferozeshah Road, New Delhi - 110 001 © ICHR & Ministry of Culture, GoI No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or any information storage and retrieval system, without permission in writing from the publisher. ISBN 978-81-938176-1-2 Printed in India by MANAK PUBLICATIONS PVT. LTD B-7, Saraswati Complex, Subhash Chowk, Laxmi Nagar, New Delhi 110092 INDIA Phone: 22453894, 22042529 [email protected] State Co-ordinators and their Researchers Andhra Pradesh & Telangana Karnataka (Co-ordinator) (Co-ordinator) V. Ramakrishna B. Surendra Rao S.K. Aruni Research Assistants Research Assistants V. Ramakrishna Reddy A.B. Vaggar I. Sudarshan Rao Ravindranath B.Venkataiah Tamil Nadu Kerala (Co-ordinator) (Co-ordinator) N. -
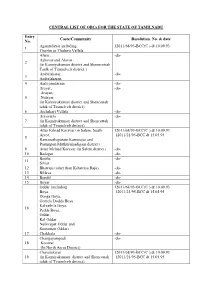
CENTRAL LIST of Obcs for the STATE of TAMILNADU Entry No
CENTRAL LIST OF OBC FOR THE STATE OF TAMILNADU E C/Cmm Rsoluti No. & da N. Agamudayar including 12011/68/93-BCC(C ) dt 10.09.93 1 Thozhu or Thuluva Vellala Alwar, -do- Azhavar and Alavar 2 (in Kanniyakumari district and Sheoncottah Taulk of Tirunelveli district ) Ambalakarar, -do- 3 Ambalakaran 4 Andi pandaram -do- Arayar, -do- Arayan, 5 Nulayar (in Kanniyakumari district and Shencottah taluk of Tirunelveli district) 6 Archakari Vellala -do- Aryavathi -do- 7 (in Kanniyakumari district and Shencottah taluk of Tirunelveli district) Attur Kilnad Koravar (in Salem, South 12011/68/93-BCC(C ) dt 10.09.93 Arcot, 12011/21/95-BCC dt 15.05.95 8 Ramanathapuram Kamarajar and Pasumpon Muthuramadigam district) 9 Attur Melnad Koravar (in Salem district) -do- 10 Badagar -do- Bestha -do- 11 Siviar 12 Bhatraju (other than Kshatriya Raju) -do- 13 Billava -do- 14 Bondil -do- 15 Boyar -do- Oddar (including 12011/68/93-BCC(C ) dt 10.09.93 Boya, 12011/21/95-BCC dt 15.05.95 Donga Boya, Gorrela Dodda Boya Kalvathila Boya, 16 Pedda Boya, Oddar, Kal Oddar Nellorepet Oddar and Sooramari Oddar) 17 Chakkala -do- Changayampadi -do- 18 Koravar (In North Arcot District) Chavalakarar 12011/68/93-BCC(C ) dt 10.09.93 19 (in Kanniyakumari district and Shencottah 12011/21/95-BCC dt 15.05.95 taluk of Tirunelveli district) Chettu or Chetty (including 12011/68/93-BCC(C ) dt 10.09.93 Kottar Chetty, 12011/21/95-BCC dt 15.05.95 Elur Chetty, Pathira Chetty 20 Valayal Chetty Pudukkadai Chetty) (in Kanniyakumari district and Shencottah taluk of Tirunelveli district) C.K. -

Human Mtdna Hypervariable Regions, HVR I and II, Hint at Deep Common Maternal Founder and Subsequent Maternal Gene flow in Indian Population Groups
J Hum Genet (2005) 50: 497–506 DOI 10.1007/s10038-005-0284-2 ORIGINAL ARTICLE Swarkar Sharma Æ Anjana Saha Æ Ekta Rai Audesh Bhat Æ Ramesh Bamezai Human mtDNA hypervariable regions, HVR I and II, hint at deep common maternal founder and subsequent maternal gene flow in Indian population groups Received: 6 May 2005 / Accepted: 21 July 2005 / Published online: 5 October 2005 Ó The Japan Society of Human Genetics and Springer-Verlag 2005 Abstract We have analysed the hypervariable regions diversity in human populations is low relative to that in (HVR I and II) of human mitochondrial DNA many other species, attesting to the recent origin and (mtDNA) in individuals from Uttar Pradesh (UP), Bihar small size of the ancestral human population (Li and (BI) and Punjab (PUNJ), belonging to the Indo-Euro- Sadler 1991; Crouau-Roy et al 1996; Kaessmann et al pean linguistic group, and from South India (SI), that 1999). Since the seminal study of Cann et al (1987), have their linguistic roots in Dravidian language. Our mitochondrial DNA (mtDNA) data have proven to be analysis revealed the presence of known and novel extremely useful for studying human evolution, includ- mutations in both hypervariable regions in the studied ing prehistoric migrations and demographic events such population groups. Median joining network analyses as sudden population expansions or extreme bottlenecks based on mtDNA showed extensive overlap in mtDNA (Sherry et al 1994). lineages despite the extensive cultural and linguistic The human mtDNA is a closed circular genome of diversity. MDS plot analysis based on Fst distances 16.5 kb in length (Anderson et al 1981), which includes suggested increased maternal genetic proximity for the a 1.1 kb-long noncoding (control) region that represents studied population groups compared with other world a highly variable sequence (Greenberg et al 1983; Melton populations. -

Editors Seek the Blessings of Mahasaraswathi
OM GAM GANAPATHAYE NAMAH I MAHASARASWATHYAI NAMAH Editors seek the blessings of MahaSaraswathi Kamala Shankar (Editor-in-Chief) Laxmikant Joshi Chitra Padmanabhan Madhu Ramesh Padma Chari Arjun I Shankar Srikali Varanasi Haranath Gnana Varsha Narasimhan II Thanks to the Authors Adarsh Ravikumar Omsri Bharat Akshay Ravikumar Prerana Gundu Ashwin Mohan Priyanka Saha Anand Kanakam Pranav Raja Arvind Chari Pratap Prasad Aravind Rajagopalan Pavan Kumar Jonnalagadda Ashneel K Reddy Rohit Ramachandran Chandrashekhar Suresh Rohan Jonnalagadda Divya Lambah Samika S Kikkeri Divya Santhanam Shreesha Suresha Dr. Dharwar Achar Srinivasan Venkatachari Girish Kowligi Srinivas Pyda Gokul Kowligi Sahana Kribakaran Gopi Krishna Sruti Bharat Guruganesh Kotta Sumedh Goutam Vedanthi Harsha Koneru Srinath Nandakumar Hamsa Ramesha Sanjana Srinivas HCCC Y&E Balajyothi class S Srinivasan Kapil Gururangan Saurabh Karmarkar Karthik Gururangan Sneha Koneru Komal Sharma Sadhika Malladi Katyayini Satya Srivishnu Goutam Vedanthi Kaushik Amancherla Saransh Gupta Medha Raman Varsha Narasimhan Mahadeva Iyer Vaishnavi Jonnalagadda M L Swamy Vyleen Maheshwari Reddy Mahith Amancherla Varun Mahadevan Nikky Cherukuthota Vaishnavi Kashyap Narasimham Garudadri III Contents Forword VI Preface VIII Chairman’s Message X President’s Message XI Significance of Maha Kumbhabhishekam XII Acharya Bharadwaja 1 Acharya Kapil 3 Adi Shankara 6 Aryabhatta 9 Bhadrachala Ramadas 11 Bhaskaracharya 13 Bheeshma 15 Brahmagupta Bhillamalacarya 17 Chanakya 19 Charaka 21 Dhruva 25 Draupadi 27 Gargi