Assessing Various Control Samples for Microarray Gene Expression Profiling of Laryngeal Squamous Cell Carcinoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Single-Cell Rnaseq Reveals Seven Classes of Colonic Sensory Neuron
Gut Online First, published on February 26, 2018 as 10.1136/gutjnl-2017-315631 Neurogastroenterology ORIGINAL ARTICLE Gut: first published as 10.1136/gutjnl-2017-315631 on 26 February 2018. Downloaded from Single-cell RNAseq reveals seven classes of colonic sensory neuron James R F Hockley,1,2 Toni S Taylor,1 Gerard Callejo,1 Anna L Wilbrey,2 Alex Gutteridge,2 Karsten Bach,1 Wendy J Winchester,2 David C Bulmer,1 Gordon McMurray,2 Ewan St John Smith1 ► Additional material is ABSTRact pathways to the central nervous system (CNS).1 In published online only. To view Objective Integration of nutritional, microbial and the colorectum, sensory innervation is organised please visit the journal online (http:// dx. doi. org/ 10. 1136/ inflammatory events along the gut-brain axis can alter into two main pathways: thoracolumbar (TL) spinal gutjnl- 2017- 315631). bowel physiology and organism behaviour. Colonic afferents projecting via the lumbar splanchnic sensory neurons activate reflex pathways and give nerve (LSN) and lumbosacral (LS) spinal afferents 1Department of Pharmacology, University of Cambridge, rise to conscious sensation, but the diversity and projecting via the pelvic nerve (PN) that are respon- Cambridge, UK division of function within these neurons is poorly sible for transducing conscious sensations of full- 2Neuroscience and Pain understood. The identification of signalling pathways ness, discomfort, urgency and pain, in addition to Research Unit, Pfizer, contributing to visceral sensation is constrained by a reflex actions.2 Cambridge, UK paucity of molecular markers. Here we address this by Visceral sensory afferents act to maintain many comprehensive transcriptomic profiling and unsupervised aspects of GI physiology, such as continence and Correspondence to James R F Hockley, Department clustering of individual mouse colonic sensory neurons. -

F2RL2 Antibody Cat
F2RL2 Antibody Cat. No.: 56-323 F2RL2 Antibody F2RL2 Antibody immunohistochemistry analysis in formalin fixed and paraffin embedded human heart tissue followed by peroxidase conjugation of the secondary antibody and DAB staining. Specifications HOST SPECIES: Rabbit SPECIES REACTIVITY: Human This F2RL2 antibody is generated from rabbits immunized with a KLH conjugated IMMUNOGEN: synthetic peptide between 21-50 amino acids from the N-terminal region of human F2RL2. TESTED APPLICATIONS: IHC-P, WB For WB starting dilution is: 1:1000 APPLICATIONS: For IHC-P starting dilution is: 1:10~50 PREDICTED MOLECULAR 43 kDa WEIGHT: September 25, 2021 1 https://www.prosci-inc.com/f2rl2-antibody-56-323.html Properties This antibody is purified through a protein A column, followed by peptide affinity PURIFICATION: purification. CLONALITY: Polyclonal ISOTYPE: Rabbit Ig CONJUGATE: Unconjugated PHYSICAL STATE: Liquid BUFFER: Supplied in PBS with 0.09% (W/V) sodium azide. CONCENTRATION: batch dependent Store at 4˚C for three months and -20˚C, stable for up to one year. As with all antibodies STORAGE CONDITIONS: care should be taken to avoid repeated freeze thaw cycles. Antibodies should not be exposed to prolonged high temperatures. Additional Info OFFICIAL SYMBOL: F2RL2 Proteinase-activated receptor 3, PAR-3, Coagulation factor II receptor-like 2, Thrombin ALTERNATE NAMES: receptor-like 2, F2RL2, PAR3 ACCESSION NO.: O00254 GENE ID: 2151 USER NOTE: Optimal dilutions for each application to be determined by the researcher. Background and References Coagulation factor II (thrombin) receptor-like 2 (F2RL2) is a member of the large family of 7-transmembrane-region receptors that couple to guanosine-nucleotide-binding proteins. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -
![Cd1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719](https://docslib.b-cdn.net/cover/1446/cd1a-o10-concentrated-and-prediluted-monoclonal-antibody-901-3158-061719-341446.webp)
Cd1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719
CD1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719 Catalog Number: ACI 3158 A, B API 3158 AA VLTM 3158 G20 Description: 0.1, 0.5 mL conc. 6.0 mL, RTU 20 mL, RTU Dilution: 1:100 Ready-to-use Ready-to-use Diluent: Van Gogh Yellow N/A N/A Intended Use: Protocol Recommendations (VALENT® Automated Slide For In Vitro Diagnostic Use Staining Platform) Cont’d: CD1a [O10] is a mouse monoclonal antibody that is intended for Protein Block (Optional): Incubate for 10-20 minutes at RT with Val laboratory use in the qualitative identification of CD1a protein by Background Block. immunohistochemistry (IHC) in formalin-fixed paraffin-embedded Primary Antibody: Incubate for 30 minutes. (FFPE) human tissues. The clinical interpretation of any staining or its Secondary: Incubate for 10 minutes with Val Mouse Secondary. absence should be complemented by morphological studies using proper Linker: Incubate for 10 minutes with Val Universal Linker. controls and should be evaluated within the context of the patient’s Polymer: Incubate for 10 minutes with Val Universal Polymer. clinical history and other diagnostic tests by a qualified pathologist. Chromogen: Incubate for 5 minutes with Val DAB. Summary and Explanation: Counterstain: Counterstain for 5 minutes with Val Hematoxylin. CD1a is a protein of 43 to 49 kDa and is expressed on dendritic cells and cortical thymocytes (1,2). CD1a [O10] staining has been shown to be Protocol Recommendations (intelliPATH FLX® and manual use): useful in the differentiation of Langerhans cells from interdigitating cells. Peroxide Block: Block for 5 minutes with Peroxidazed 1. It has also proved useful for phenotyping Langerhans cell histiocytosis Pretreatment: Perform heat retrieval using Diva or Reveal Decloaker. -

Monitoring Nociception by Analyzing Gene Expression Changes in the Central Nervous System of Mice
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2010 Monitoring nociception by analyzing gene expression changes in the central nervous system of mice Asner, I N Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-46678 Dissertation Originally published at: Asner, I N. Monitoring nociception by analyzing gene expression changes in the central nervous system of mice. 2010, University of Zurich, Vetsuisse Faculty. Monitoring Nociception by Analyzing Gene Expression Changes in the Central Nervous System of Mice Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr. sc. nat) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultät der Universität Zürich von Igor Asner von St. Cergue VD Promotionskomitee Prof. Dr. Peter Sonderegger Prof. Dr. Kurt Bürki Prof. Dr. Hanns Ulrich Zeilhofer Dr. Paolo Cinelli (Leitung der Dissertation) Zürich, 2010 Table of contents Table of content Curriculum vitae 6 Publications 9 Summary 11 Zusammenfassung 14 1. Introduction 17 1.1. Pain and nociception 17 1.1.1 Nociceptive neurons and Mechanoceptors 18 1.1.2 Activation of the nociceptive neurons at the periphery 21 1.1.2.1 Response to noxious heat 22 1.1.2.2 Response to noxious cold 23 1.1.2.3 Response to mechanical stress 24 1.1.3 Nociceptive message processing in the Spinal Cord 25 1.1.3.1 The lamina I and the ascending pathways 25 1.1.3.2 The lamina II and the descending pathways 26 1.1.4 Pain processing and integration in the brain 27 1.1.4.1 The Pain Matrix 27 1.1.4.2 Activation of the descending pathways 29 1.1.5 Inflammatory Pain 31 1.2. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -

Integrative Differential Expression and Gene Set Enrichment Analysis Using Summary Statistics for Scrna-Seq Studies
ARTICLE https://doi.org/10.1038/s41467-020-15298-6 OPEN Integrative differential expression and gene set enrichment analysis using summary statistics for scRNA-seq studies ✉ Ying Ma 1,7, Shiquan Sun 1,7, Xuequn Shang2, Evan T. Keller 3, Mengjie Chen 4,5 & Xiang Zhou 1,6 Differential expression (DE) analysis and gene set enrichment (GSE) analysis are commonly applied in single cell RNA sequencing (scRNA-seq) studies. Here, we develop an integrative 1234567890():,; and scalable computational method, iDEA, to perform joint DE and GSE analysis through a hierarchical Bayesian framework. By integrating DE and GSE analyses, iDEA can improve the power and consistency of DE analysis and the accuracy of GSE analysis. Importantly, iDEA uses only DE summary statistics as input, enabling effective data modeling through com- plementing and pairing with various existing DE methods. We illustrate the benefits of iDEA with extensive simulations. We also apply iDEA to analyze three scRNA-seq data sets, where iDEA achieves up to five-fold power gain over existing GSE methods and up to 64% power gain over existing DE methods. The power gain brought by iDEA allows us to identify many pathways that would not be identified by existing approaches in these data. 1 Department of Biostatistics, University of Michigan, Ann Arbor, MI 48109, USA. 2 School of Computer Science, Northwestern Polytechnical University, Xi’an, Shaanxi 710072, P.R. China. 3 Department of Urology, University of Michigan, Ann Arbor, MI 48109, USA. 4 Department of Human Genetics, University of Chicago, Chicago, IL 60637, USA. 5 Section of Genetic Medicine, Department of Medicine, University of Chicago, Chicago, IL 60637, USA. -
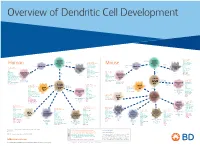
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

Gene List HTG Edgeseq Immuno-Oncology Assay
Gene List HTG EdgeSeq Immuno-Oncology Assay Adhesion ADGRE5 CLEC4A CLEC7A IBSP ICAM4 ITGA5 ITGB1 L1CAM MBL2 SELE ALCAM CLEC4C DST ICAM1 ITGA1 ITGA6 ITGB2 LGALS1 MUC1 SVIL CDH1 CLEC5A EPCAM ICAM2 ITGA2 ITGAL ITGB3 LGALS3 NCAM1 THBS1 CDH5 CLEC6A FN1 ICAM3 ITGA4 ITGAM ITGB4 LGALS9 PVR THY1 Apoptosis APAF1 BCL2 BID CARD11 CASP10 CASP8 FADD NOD1 SSX1 TP53 TRAF3 BCL10 BCL2L1 BIRC5 CASP1 CASP3 DDX58 NLRP3 NOD2 TIMP1 TRAF2 TRAF6 B-Cell Function BLNK BTLA CD22 CD79A FAS FCER2 IKBKG PAX5 SLAMF1 SLAMF7 SPN BTK CD19 CD24 EBF4 FASLG IKBKB MS4A1 RAG1 SLAMF6 SPI1 Cell Cycle ABL1 ATF1 ATM BATF CCND1 CDK1 CDKN1B NCL RELA SSX1 TBX21 TP53 ABL2 ATF2 AXL BAX CCND3 CDKN1A EGR1 REL RELB TBK1 TIMP1 TTK Cell Signaling ADORA2A DUSP4 HES1 IGF2R LYN MAPK1 MUC1 NOTCH1 RIPK2 SMAD3 STAT5B AKT3 DUSP6 HES5 IKZF1 MAF MAPK11 MYC PIK3CD RNF4 SOCS1 STAT6 BCL6 ELK1 HEY1 IKZF2 MAP2K1 MAPK14 NFATC1 PIK3CG RORC SOCS3 SYK CEBPB EP300 HEY2 IKZF3 MAP2K2 MAPK3 NFATC3 POU2F2 RUNX1 SPINK5 TAL1 CIITA ETS1 HEYL JAK1 MAP2K4 MAPK8 NFATC4 PRKCD RUNX3 STAT1 TCF7 CREB1 FLT3 HMGB1 JAK2 MAP2K7 MAPKAPK2 NFKB1 PRKCE S100B STAT2 TYK2 CREB5 FOS HRAS JAK3 MAP3K1 MEF2C NFKB2 PTEN SEMA4D STAT3 CREBBP GATA3 IGF1R KIT MAP3K5 MTDH NFKBIA PYCARD SMAD2 STAT4 Chemokine CCL1 CCL16 CCL20 CCL25 CCL4 CCR2 CCR7 CX3CL1 CXCL12 CXCL3 CXCR1 CXCR6 CCL11 CCL17 CCL21 CCL26 CCL5 CCR3 CCR9 CX3CR1 CXCL13 CXCL5 CXCR2 MST1R CCL13 CCL18 CCL22 CCL27 CCL7 CCR4 CCRL2 CXCL1 CXCL14 CXCL6 CXCR3 PPBP CCL14 CCL19 CCL23 CCL28 CCL8 CCR5 CKLF CXCL10 CXCL16 CXCL8 CXCR4 XCL2 CCL15 CCL2 CCL24 CCL3 CCR1 CCR6 CMKLR1 CXCL11 CXCL2 CXCL9 CXCR5