Open Research Online Oro.Open.Ac.Uk
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RSC Advances
RSC Advances This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. This Accepted Manuscript will be replaced by the edited, formatted and paginated article as soon as this is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/advances Page 1 of 8 RSC Advances The Effects of Solvent Composition on the Affinity of a Peptide towards Hair Keratin: Experimental and Molecular Dynamics Data. Egipto Antunes a, Célia F. Cruz a, Nuno G. Azoia a, Artur Cavaco-Paulo a* a CEB – Centre of Biological Engineering, University of Minho, 4710-057 Braga, Portugal Table of Contents Manuscript Accepted Molecular dynamics simulations with a developed hair protofibril model demonstrated capability to improve a peptide uptake by hair shafts. Advances RSC RSC Advances Page 2 of 8 RSC Advances RSCPublishing ARTICLE The Effects of Solvent Composition on the Affinity of a Peptide towards Hair Keratin: Experimental and Cite this: DOI: 10.1039/x0xx00000x Molecular Dynamics Data. -

MALE Protein Name Accession Number Molecular Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean H Mean PDAC Mean T-Test PDAC Vs. H T-Test
MALE t-test t-test Accession Molecular H PDAC PDAC vs. PDAC vs. Protein Name Number Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean Mean Mean H CP PDAC/H PDAC/CP - 22 kDa protein IPI00219910 22 kDa 7 5 4 8 1 0 6 6 1 0.1126 0.0456 0.1 0.1 - Cold agglutinin FS-1 L-chain (Fragment) IPI00827773 12 kDa 32 39 34 26 53 57 36 30 55 0.0309 0.0388 1.8 1.5 - HRV Fab 027-VL (Fragment) IPI00827643 12 kDa 4 6 0 0 0 0 5 0 0 - 0.0574 - 0.0 - REV25-2 (Fragment) IPI00816794 15 kDa 8 12 5 7 8 9 10 6 8 0.2225 0.3844 1.3 0.8 A1BG Alpha-1B-glycoprotein precursor IPI00022895 54 kDa 115 109 106 112 111 100 112 109 105 0.6497 0.4138 1.0 0.9 A2M Alpha-2-macroglobulin precursor IPI00478003 163 kDa 62 63 86 72 14 18 63 79 16 0.0120 0.0019 0.2 0.3 ABCB1 Multidrug resistance protein 1 IPI00027481 141 kDa 41 46 23 26 52 64 43 25 58 0.0355 0.1660 2.4 1.3 ABHD14B Isoform 1 of Abhydrolase domain-containing proteinIPI00063827 14B 22 kDa 19 15 19 17 15 9 17 18 12 0.2502 0.3306 0.7 0.7 ABP1 Isoform 1 of Amiloride-sensitive amine oxidase [copper-containing]IPI00020982 precursor85 kDa 1 5 8 8 0 0 3 8 0 0.0001 0.2445 0.0 0.0 ACAN aggrecan isoform 2 precursor IPI00027377 250 kDa 38 30 17 28 34 24 34 22 29 0.4877 0.5109 1.3 0.8 ACE Isoform Somatic-1 of Angiotensin-converting enzyme, somaticIPI00437751 isoform precursor150 kDa 48 34 67 56 28 38 41 61 33 0.0600 0.4301 0.5 0.8 ACE2 Isoform 1 of Angiotensin-converting enzyme 2 precursorIPI00465187 92 kDa 11 16 20 30 4 5 13 25 5 0.0557 0.0847 0.2 0.4 ACO1 Cytoplasmic aconitate hydratase IPI00008485 98 kDa 2 2 0 0 0 0 2 0 0 - 0.0081 - 0.0 -

Pflugers Final
CORE Metadata, citation and similar papers at core.ac.uk Provided by Serveur académique lausannois A comprehensive analysis of gene expression profiles in distal parts of the mouse renal tubule. Sylvain Pradervand2, Annie Mercier Zuber1, Gabriel Centeno1, Olivier Bonny1,3,4 and Dmitri Firsov1,4 1 - Department of Pharmacology and Toxicology, University of Lausanne, 1005 Lausanne, Switzerland 2 - DNA Array Facility, University of Lausanne, 1015 Lausanne, Switzerland 3 - Service of Nephrology, Lausanne University Hospital, 1005 Lausanne, Switzerland 4 – these two authors have equally contributed to the study to whom correspondence should be addressed: Dmitri FIRSOV Department of Pharmacology and Toxicology, University of Lausanne, 27 rue du Bugnon, 1005 Lausanne, Switzerland Phone: ++ 41-216925406 Fax: ++ 41-216925355 e-mail: [email protected] and Olivier BONNY Department of Pharmacology and Toxicology, University of Lausanne, 27 rue du Bugnon, 1005 Lausanne, Switzerland Phone: ++ 41-216925417 Fax: ++ 41-216925355 e-mail: [email protected] 1 Abstract The distal parts of the renal tubule play a critical role in maintaining homeostasis of extracellular fluids. In this review, we present an in-depth analysis of microarray-based gene expression profiles available for microdissected mouse distal nephron segments, i.e., the distal convoluted tubule (DCT) and the connecting tubule (CNT), and for the cortical portion of the collecting duct (CCD) (Zuber et al., 2009). Classification of expressed transcripts in 14 major functional gene categories demonstrated that all principal proteins involved in maintaining of salt and water balance are represented by highly abundant transcripts. However, a significant number of transcripts belonging, for instance, to categories of G protein-coupled receptors (GPCR) or serine-threonine kinases exhibit high expression levels but remain unassigned to a specific renal function. -

University of Alberta
University of Alberta T cell-mediated inflammation is stereotyped: mouse delayed-type hypersensitivity reaction and mouse T cell-mediated rejection of renal allografts share common molecular mechanisms by Jeffery M. Venner A thesis submitted to the Faculty of Graduate Studies and Research in partial fulfillment of the requirements for the degree of Master of Science in Experimental Medicine Department of Medicine ©Jeffery M. Venner Fall 2011 Edmonton, Alberta Permission is hereby granted to the University of Alberta Libraries to reproduce single copies of this thesis and to lend or sell such copies for private, scholarly or scientific research purposes only. Where the thesis is converted to, or otherwise made available in digital form, the University of Alberta will advise potential users of the thesis of these terms. The author reserves all other publication and other rights in association with the copyright in the thesis and, except as herein before provided, neither the thesis nor any substantial portion thereof may be printed or otherwise reproduced in any material form whatsoever without the author's prior written permission. Dedicated to those who have inspired me: My family and friends, my teachers and mentors. You laid the foundation – my success is guaranteed ABSTRACT Genome-wide gene expression analysis of diseases has revealed large-scale changes in the expression of thousands of genes (transcripts) representing biological processes. The processes that occur during T cell-mediated rejection (TCMR) of renal allografts in mice and humans have been previously delineated, and they appear to be independent of cytotoxic mechanisms; thus, TCMR is analogous to delayed-type hypersensitivity (DTH). -

Transcriptome Profiling and Differential Gene Expression In
G C A T T A C G G C A T genes Article Transcriptome Profiling and Differential Gene Expression in Canine Microdissected Anagen and Telogen Hair Follicles and Interfollicular Epidermis Dominique J. Wiener 1,* ,Kátia R. Groch 1 , Magdalena A.T. Brunner 2,3, Tosso Leeb 2,3 , Vidhya Jagannathan 2 and Monika M. Welle 3,4 1 Department of Veterinary Pathobiology, College of Veterinary Medicine & Biomedical Science, Texas A&M University, College Station, TX 77843, USA; [email protected] 2 Institute of Genetics, Vetsuisse Faculty, University of Bern, 3012 Bern, Switzerland; [email protected] (M.A.T.B.); [email protected] (T.L.); [email protected] (V.J.) 3 Dermfocus, Vetsuisse Faculty, University Hospital of Bern, 3010 Bern, Switzerland; [email protected] 4 Institute of Animal Pathology, Vetsuisse Faculty, University of Bern, 3012 Bern, Switzerland * Correspondence: [email protected]; Tel.: +1-979-862-1568 Received: 30 June 2020; Accepted: 3 August 2020; Published: 4 August 2020 Abstract: The transcriptome profile and differential gene expression in telogen and late anagen microdissected hair follicles and the interfollicular epidermis of healthy dogs was investigated by using RNAseq. The genes with the highest expression levels in each group were identified and genes known from studies in other species to be associated with structure and function of hair follicles and epidermis were evaluated. Transcriptome profiling revealed that late anagen follicles expressed mainly keratins and telogen follicles expressed GSN and KRT15. The interfollicular epidermis expressed predominately genes encoding for proteins associated with differentiation. All sample groups express genes encoding for proteins involved in cellular growth and signal transduction. -

Technology for Proteomics Translation to Clinical Research
TechnologyTechnology ForFor ProteomicsProteomics Translation to Clinical ResearchResearch StudiesStudies Lance A. Liotta MD PhD George Mason University A. Novel one step preservative for tissue phosphoproteins B. Protein Microarrays: 200 signal pathway phosphoproteins -Translation to clinical research trials -The universal tissue preservative: obviate frozen storage C. Smart nanoparticles for one step in-solution molecular size sieving, affinity capture, biomarker preservation and amplification of effective sensitivity. The Center for Applied Proteomics and M ole cular Medicine Proteomics Tools for Clinical Medicine There is a need to measure the state of activity of the actual drug targets (the proteins) in a patient’s individual cancer. Patient A Patient B Proteomics is the missing link for designing individualized therapies Concurrent phosphorylation of kinases and kinase substrates indicates functional linkage “Proteins carry the epigenetic marks and information” Emma Whitelaw, DISCOVER Nov. 2006 Genetic or epigenetic defects are selected during cancer progression because they cooperate to orchestrate alterations in protein networks generating a survival advantage for the target cell. Post-translational modifications, such as phosphorylation, reflect the activity state of cellular signaling networks. Patterns of phosphorylation indicate docking events and infer protein-protein interactions. Pre‐analytical Variables: The tissue is alive! •The tissue is alive and reactive post excision • During the post excision delay time, tissue signal -

Keratin-Mediated Hair Growth
Keratin is not only a Structural Protein in Hair: Keratin-mediated Hair Growth Seong Yeong An Kyung Hee University Eun Ji Choi Konkuk University So Yeon Kim Kyung Hee University Se Young Van Kyung Hee University Han Jun Kim Konkuk University Jae-Hyung Lee Kyung Hee University https://orcid.org/0000-0002-5085-6988 Song Wook Han KeraMedix Inc Il Keun Kwon Kyung Hee University Chul-Kyu Lee Chemon Inc. Sun Hee Do Konkuk University Yu-Shik Hwang ( [email protected] ) Kyung Hee University Article Keywords: Keratin , hair, intradermal injection, outer root sheath cells, Posted Date: November 18th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-101358/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/27 Abstract Keratin is known to be a major protein in hair, but the biological function of keratin in hair growth is unknown, which led us to conduct a pilot study to elucidate biological function of keratin in hair growth via cellular interactions with hair forming cells. Here, we show hair growth is stimulated by intradermal injection of keratin into mice, and show that outer root sheath cells undergo transforming growth factor- β2-induced apoptosis, resulting in keratin exposure. Keratin exposure appears to be critical for dermal papilla cell condensation and hair germ formation as immunodepletion and silencing keratin prevent dermal papilla cell condensation and hair germ formation. Furthermore, silencing keratin in mice resulted in a marked suppression of anagen follicle formation and hair growth. Our study imply a new nding of how to initiate hair regeneration and suggests the potent application of keratin biomaterial for the treatment of hair loss. -
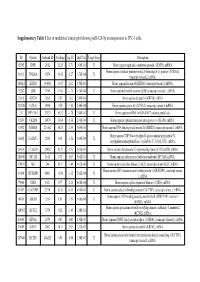
Supplementary Table 1 List of Modulated Transcripts Following Mir-129-5P Overexpression in TPC-1 Cells
Supplementary Table 1 List of modulated transcripts following miR-129-5p overexpression in TPC-1 cells. ID Symbol Genbank ID AveExpr log_FC adj.P.Val Target Scan Description 92585 EMP1 2012 12.15 -3.71 1.06E-04 X Homo sapiens epithelial membrane protein 1 (EMP1), mRNA. Homo sapiens vacuolar protein sorting 26 homolog A (S. pombe) (VPS26A), 10111 VPS26A 9559 11.92 -2.27 1.70E-04 X transcript variant 2, mRNA. 186912 KRT80 144501 11.97 2.62 1.70E-04 Homo sapiens keratin 80 (KRT80), transcript variant 2, mRNA. 92202 LBR 3930 11.86 -1.75 1.70E-04 X Homo sapiens lamin B receptor (LBR), transcript variant 1, mRNA. 33816 KRT34 3885 7.87 1.62 2.48E-04 Homo sapiens keratin 34 (KRT34), mRNA. 121520 CCNA1 8900 9.50 -1.80 2.48E-04 Homo sapiens cyclin A1 (CCNA1), transcript variant 4, mRNA. 211 DPY19L1 23333 10.32 -1.78 2.48E-04 X Homo sapiens mRNA for KIAA0877 protein, partial cds. 15854 CKAP4 10970 13.04 -1.36 2.66E-04 X Homo sapiens cytoskeleton-associated protein 4 (CKAP4), mRNA. 63392 RBM24 221662 10.83 2.39 3.09E-04 Homo sapiens RNA binding motif protein 24 (RBM24), transcript variant 2, mRNA. Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N- 36665 GALNT1 2589 9.61 -1.52 3.09E-04 X acetylgalactosaminyltransferase 1 (GalNAc-T1) (GALNT1), mRNA. 18614 C12orf24 29902 10.57 1.76 5.02E-04 Homo sapiens chromosome 12 open reading frame 24 (C12orf24), mRNA. 126848 MCAM 4162 8.72 2.05 5.02E-04 X Homo sapiens melanoma cell adhesion molecule (MCAM), mRNA. -

1 SUPPLEMENTAL DATA Figure S1. Poly I:C Induces IFN-Β Expression
SUPPLEMENTAL DATA Figure S1. Poly I:C induces IFN-β expression and signaling. Fibroblasts were incubated in media with or without Poly I:C for 24 h. RNA was isolated and processed for microarray analysis. Genes showing >2-fold up- or down-regulation compared to control fibroblasts were analyzed using Ingenuity Pathway Analysis Software (Red color, up-regulation; Green color, down-regulation). The transcripts with known gene identifiers (HUGO gene symbols) were entered into the Ingenuity Pathways Knowledge Base IPA 4.0. Each gene identifier mapped in the Ingenuity Pathways Knowledge Base was termed as a focus gene, which was overlaid into a global molecular network established from the information in the Ingenuity Pathways Knowledge Base. Each network contained a maximum of 35 focus genes. 1 Figure S2. The overlap of genes regulated by Poly I:C and by IFN. Bioinformatics analysis was conducted to generate a list of 2003 genes showing >2 fold up or down- regulation in fibroblasts treated with Poly I:C for 24 h. The overlap of this gene set with the 117 skin gene IFN Core Signature comprised of datasets of skin cells stimulated by IFN (Wong et al, 2012) was generated using Microsoft Excel. 2 Symbol Description polyIC 24h IFN 24h CXCL10 chemokine (C-X-C motif) ligand 10 129 7.14 CCL5 chemokine (C-C motif) ligand 5 118 1.12 CCL5 chemokine (C-C motif) ligand 5 115 1.01 OASL 2'-5'-oligoadenylate synthetase-like 83.3 9.52 CCL8 chemokine (C-C motif) ligand 8 78.5 3.25 IDO1 indoleamine 2,3-dioxygenase 1 76.3 3.5 IFI27 interferon, alpha-inducible -

Involvement of TGF-ß Receptor– and Integrin-Mediated Signaling Pathways in the Pathogenesis of Granular Corneal Dystrophy II
Biochemistry and Molecular Biology Involvement of TGF- Receptor– and Integrin-Mediated Signaling Pathways in the Pathogenesis of Granular Corneal Dystrophy II Seung-il Choi,1 Yeong-Min Yoo,2 Bong-Yoon Kim,1 Tae-im Kim,1 Hyun-ju Cho,1 So-yoen Ahn,1 Hyung Keun Lee,1 Hyun-Soo Cho,3 and Eung Kweon Kim1 PURPOSE. The purpose of this study was to elucidate the (Invest Ophthalmol Vis Sci. 2010;51:1832–1847) DOI: pathophysiological process in primary cultured corneal fibro- 10.1167/iovs.09-4149 blasts (PCFs) from normal subjects and granular corneal dys- trophy (GCD) II patients, by using cDNA microarrays. ranular corneal dystrophy II (GCD II) is a disorder char- METHODS. PCFs were isolated from the corneas of normal Gacterized by age-dependent progressive accumulation of subjects and GCD II patients who were heterozygous and protein deposits in the corneal epithelia and stroma, followed homozygous for the TGFBI R124H mutation. RNA was isolated by disruption of corneal transparency. GCD II is an autosomal from each sample, and gene expression profiles were analyzed dominant disorder caused by a point mutation (R124H) in the with a cDNA microarray consisting of approximately 29,000 transforming growth factor--induced gene (TGFBI) on chro- genes. Cell adhesion assays were performed to confirm the mosome 5, region q31.1,2 TGFBI encodes a highly conserved functionality of the detected gene expression profiles. 683 amino acid protein (TGFBIp) that contains a secretary signal sequence and an Arg-Gly-Asp (RGD) motif that serves as RESULTS. Twofold differences were detected in the expression a ligand recognition site for integrins.1 TGFBIp is a component of 555 genes between wild-type and homozygous GCD II PCFs. -

Keratin 34 Sirna (M): Sc-146413
SANTA CRUZ BIOTECHNOLOGY, INC. Keratin 34 siRNA (m): sc-146413 BACKGROUND STORAGE AND RESUSPENSION The keratin multigene family is made of the “soft” epithelial cytokeratins and Store lyophilized siRNA duplex at -20° C with desiccant. Stable for at least the “hard” hair keratins. While the epithelial cytokeratins are involved in the one year from the date of shipment. Once resuspended, store at -20° C, layering and formation of epithelia, the hair keratins are responsible for cre- avoid contact with RNAses and repeated freeze thaw cycles. ating nails and hair. There are two types of hair keratins: the acidic type I hair Resuspend lyophilized siRNA duplex in 330 µl of the RNAse-free water keratin proteins and the basic/neutral type II hair keratin proteins. Keratin 34 provided. Resuspension of the siRNA duplex in 330 µl of RNAse-free water (K34), also known as HA4, hair keratin type I Ha4, KRTHA4 or KRT34, is a 436 makes a 10 µM solution in a 10 µM Tris-HCl, pH 8.0, 20 mM NaCl, 1 mM amino acid type I hair keratin belonging to the intermediate filament family. EDTA buffered solution. Expressed in the hair follicle, Keratin 34 forms a heterodimer with type II keratins to form nails and hair and is encoded by a gene located on human APPLICATIONS chromosome 17q21.2 and mouse chromosome 11 D. Keratin 34 siRNA (m) is recommended for the inhibition of Keratin 34 REFERENCES expression in mouse cells. 1. Heid, H.W., Werner, E. and Franke, W.W. 1986. The complement of native SUPPORT REAGENTS α-keratin polypeptides of hair-forming cells: a subset of eight polypeptides that differ from epithelial cytokeratins. -

Effects of Food-Derived Collagen Peptides on the Expression of Keratin and Keratin-Associated Protein Genes in the Mouse Skin
Original Paper Skin Pharmacol Physiol 2015;28:227–235 Received: September 15, 2014 DOI: 10.1159/000369830 Accepted after revision: November 8, 2014 Published online: February 14, 2015 Effects of Food-Derived Collagen Peptides on the Expression of Keratin and Keratin-Associated Protein Genes in the Mouse Skin a a a a a, e Phuong Le Vu Ryo Takatori Taku Iwamoto Yutaka Akagi Hideo Satsu a b d a, c Mamoru Totsuka Kazuhiro Chida Kenji Sato Makoto Shimizu a b c Departments of Applied Biological Chemistry and Animal Resource Sciences, The University of Tokyo, and Department d of Nutritional Science, Tokyo University of Agriculture, Tokyo , Division of Applied Life Sciences, Kyoto Prefectural e University, Kyoto , and Department of Biotechnology, Maebashi Institute of Technology, Maebashi , Japan Key Words were cultured without fibroblasts, suggesting that the pres- Coculture · Collagen · Food-derived collagen peptides · ence of fibroblasts is essential for the effects of Pro-Hyp. Our Gene expression · Keratinocytes · Skin study presents new insights into the effects of CP on the skin, which might link to the hair cycle. © 2015 S. Karger AG, Basel Abstract Oral ingestion of collagen peptides (CP) has long been sug- gested to exert beneficial effects on the skin, but the mo- Introduction lecular events induced by CP on the skin remain unclear. Here, we investigated the effects of oral CP administration The skin is the foremost protective barrier of the body, on gene expression in hairless mouse skin and of prolyl-hy- which acts to prevent environmental harmful factors as droxyproline (Pro-Hyp), a collagen-derived dipeptide, on well as water loss.