Bootstrapping Regression Models
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Synthpop: Bespoke Creation of Synthetic Data in R
synthpop: Bespoke Creation of Synthetic Data in R Beata Nowok Gillian M Raab Chris Dibben University of Edinburgh University of Edinburgh University of Edinburgh Abstract In many contexts, confidentiality constraints severely restrict access to unique and valuable microdata. Synthetic data which mimic the original observed data and preserve the relationships between variables but do not contain any disclosive records are one possible solution to this problem. The synthpop package for R, introduced in this paper, provides routines to generate synthetic versions of original data sets. We describe the methodology and its consequences for the data characteristics. We illustrate the package features using a survey data example. Keywords: synthetic data, disclosure control, CART, R, UK Longitudinal Studies. This introduction to the R package synthpop is a slightly amended version of Nowok B, Raab GM, Dibben C (2016). synthpop: Bespoke Creation of Synthetic Data in R. Journal of Statistical Software, 74(11), 1-26. doi:10.18637/jss.v074.i11. URL https://www.jstatsoft. org/article/view/v074i11. 1. Introduction and background 1.1. Synthetic data for disclosure control National statistical agencies and other institutions gather large amounts of information about individuals and organisations. Such data can be used to understand population processes so as to inform policy and planning. The cost of such data can be considerable, both for the collectors and the subjects who provide their data. Because of confidentiality constraints and guarantees issued to data subjects the full access to such data is often restricted to the staff of the collection agencies. Traditionally, data collectors have used anonymisation along with simple perturbation methods such as aggregation, recoding, record-swapping, suppression of sensitive values or adding random noise to prevent the identification of data subjects. -

A Survey on Data Collection for Machine Learning a Big Data - AI Integration Perspective
1 A Survey on Data Collection for Machine Learning A Big Data - AI Integration Perspective Yuji Roh, Geon Heo, Steven Euijong Whang, Senior Member, IEEE Abstract—Data collection is a major bottleneck in machine learning and an active research topic in multiple communities. There are largely two reasons data collection has recently become a critical issue. First, as machine learning is becoming more widely-used, we are seeing new applications that do not necessarily have enough labeled data. Second, unlike traditional machine learning, deep learning techniques automatically generate features, which saves feature engineering costs, but in return may require larger amounts of labeled data. Interestingly, recent research in data collection comes not only from the machine learning, natural language, and computer vision communities, but also from the data management community due to the importance of handling large amounts of data. In this survey, we perform a comprehensive study of data collection from a data management point of view. Data collection largely consists of data acquisition, data labeling, and improvement of existing data or models. We provide a research landscape of these operations, provide guidelines on which technique to use when, and identify interesting research challenges. The integration of machine learning and data management for data collection is part of a larger trend of Big data and Artificial Intelligence (AI) integration and opens many opportunities for new research. Index Terms—data collection, data acquisition, data labeling, machine learning F 1 INTRODUCTION E are living in exciting times where machine learning expertise. This problem applies to any novel application that W is having a profound influence on a wide range of benefits from machine learning. -

A First Step Into the Bootstrap World
INSTITUTE FOR DEFENSE ANALYSES A First Step into the Bootstrap World Matthew Avery May 2016 Approved for public release. IDA Document NS D-5816 Log: H 16-000624 INSTITUTE FOR DEFENSE ANALYSES 4850 Mark Center Drive Alexandria, Virginia 22311-1882 The Institute for Defense Analyses is a non-profit corporation that operates three federally funded research and development centers to provide objective analyses of national security issues, particularly those requiring scientific and technical expertise, and conduct related research on other national challenges. About This Publication This briefing motivates bootstrapping through an intuitive explanation of basic statistical inference, discusses the right way to resample for bootstrapping, and uses examples from operational testing where the bootstrap approach can be applied. Bootstrapping is a powerful and flexible tool when applied appropriately. By treating the observed sample as if it were the population, the sampling distribution of statistics of interest can be generated with precision equal to Monte Carlo error. This approach is nonparametric and requires only acceptance of the observed sample as an estimate for the population. Careful resampling is used to generate the bootstrap distribution. Applications include quantifying uncertainty for availability data, quantifying uncertainty for complex distributions, and using the bootstrap for inference, such as the two-sample t-test. Copyright Notice © 2016 Institute for Defense Analyses 4850 Mark Center Drive, Alexandria, Virginia 22311-1882 -
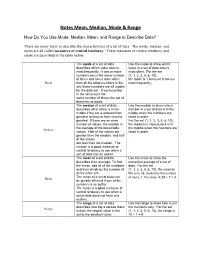
Notes Mean, Median, Mode & Range
Notes Mean, Median, Mode & Range How Do You Use Mode, Median, Mean, and Range to Describe Data? There are many ways to describe the characteristics of a set of data. The mode, median, and mean are all called measures of central tendency. These measures of central tendency and range are described in the table below. The mode of a set of data Use the mode to show which describes which value occurs value in a set of data occurs most frequently. If two or more most often. For the set numbers occur the same number {1, 1, 2, 3, 5, 6, 10}, of times and occur more often the mode is 1 because it occurs Mode than all the other numbers in the most frequently. set, those numbers are all modes for the data set. If each number in the set occurs the same number of times, the set of data has no mode. The median of a set of data Use the median to show which describes what value is in the number in a set of data is in the middle if the set is ordered from middle when the numbers are greatest to least or from least to listed in order. greatest. If there are an even For the set {1, 1, 2, 3, 5, 6, 10}, number of values, the median is the median is 3 because it is in the average of the two middle the middle when the numbers are Median values. Half of the values are listed in order. greater than the median, and half of the values are less than the median. -

Common Data Set 2020-2021 University of Pennsylvania
Common Data Set 2020-2021 University of Pennsylvania Table of Contents A General Information page 3 B Enrollment and Persistence 5 C First Time, First Year Admission 8 D Transfer Admission 14 E Academic Offerings and Policies 16 F Student Life 18 G Annual Expenses 20 H Financial Aid 22 I Instructional Faculty and Class Size 28 J Disciplinary Areas of Degrees Conferred 30 Common Data Set Definitions 32 Common Data Set 2020-21 1 25 Jun 2021 Common Data Set 2020-21 2 25 Jun 2021 A. General Information return to Table of Contents A1 Address Information A1 University of Pennsylvania A1 Mailing Address: 1 College Hall, Room 100 A1 City/State/Zip/Country: Philadelphia, PA 19104-6228 A1 Main Phone Number: 215-898-5000 A1 Home Page Address www.upenn.edu A1 Admissions Phone Number: 215-898-7507 A1 Admissions Office Mailing Address: 1 College Hall A1 City/State/Zip/Country: Philadelphia, PA 19104 A1 Admissions Fax Number: 215-898-9670 A1 Admissions E-mail Address: [email protected] A1 Online application www.admissions.upenn.edu A2 Source of institutional control (Check only one): A2 Public A2 Private (nonprofit) x A2 Proprietary A3 Classify your undergraduate institution: A3 Coeducational college A3 Men's college A3 Women's college A4 Academic year calendar: A4 Semester x A4 Quarter A4 Trimester A4 4-1-4 A4 Continuous A5 Degrees offered by your institution: A5 Certificate x A5 Diploma A5 Associate x A5 Transfer Associate A5 Terminal Associate x A5 Bachelor's x A5 Postbachelor's certificate x A5 Master's x A5 Post-master's certificate x A5 Doctoral degree - research/scholarship x A5 Doctoral degree - professional practice x A5 Doctoral degree - other x A5 Doctoral degree -- other Common Data Set 2020-21 3 25 Jun 2021 Common Data Set 2020-21 4 25 Jun 2021 B. -

Lecture 14 Testing for Kurtosis
9/8/2016 CHE384, From Data to Decisions: Measurement, Kurtosis Uncertainty, Analysis, and Modeling • For any distribution, the kurtosis (sometimes Lecture 14 called the excess kurtosis) is defined as Testing for Kurtosis 3 (old notation = ) • For a unimodal, symmetric distribution, Chris A. Mack – a positive kurtosis means “heavy tails” and a more Adjunct Associate Professor peaked center compared to a normal distribution – a negative kurtosis means “light tails” and a more spread center compared to a normal distribution http://www.lithoguru.com/scientist/statistics/ © Chris Mack, 2016Data to Decisions 1 © Chris Mack, 2016Data to Decisions 2 Kurtosis Examples One Impact of Excess Kurtosis • For the Student’s t • For a normal distribution, the sample distribution, the variance will have an expected value of s2, excess kurtosis is and a variance of 6 2 4 1 for DF > 4 ( for DF ≤ 4 the kurtosis is infinite) • For a distribution with excess kurtosis • For a uniform 2 1 1 distribution, 1 2 © Chris Mack, 2016Data to Decisions 3 © Chris Mack, 2016Data to Decisions 4 Sample Kurtosis Sample Kurtosis • For a sample of size n, the sample kurtosis is • An unbiased estimator of the sample excess 1 kurtosis is ∑ ̅ 1 3 3 1 6 1 2 3 ∑ ̅ Standard Error: • For large n, the sampling distribution of 1 24 2 1 approaches Normal with mean 0 and variance 2 1 of 24/n 3 5 • For small samples, this estimator is biased D. N. Joanes and C. A. Gill, “Comparing Measures of Sample Skewness and Kurtosis”, The Statistician, 47(1),183–189 (1998). -

Data Analysis
© Jones & Bartlett Learning, LLC. NOT FOR SALE OR DISTRIBUTION Chapter 13 Data Analysis James Eldridge © VLADGRIN/iStock/Thinkstock Chapter Objectives At the conclusion of this chapter, the learner will be able to: 1. Identify the types of statistics available for analyses in evidence-based practice. 2. Define quantitative analysis, qualitative analysis, and quality assurance. 3. Discuss how research questions define the type of statistics to be used in evidence-based practice research. 4. Choose a data analysis plan and the proper statistics for different research questions raised in evidence-based practice. 5. Interpret data analyses and conclusions from the data analyses. 6. Discuss how quality assurance affects evidence-based practice. Key Terms Analysis of variance (ANOVA) Magnitude Central tendency Mean Chi-square Median Interval scale Mode 9781284108958_CH13_Pass03.indd 375 10/17/15 10:16 AM © Jones & Bartlett Learning, LLC. NOT FOR SALE OR DISTRIBUTION 376 | Chapter 13 Data Analysis Nominal scale Quantitative analysis Ordinal scale Ratio scale Qualitative analysis Statistical Package for the Social Quality assurance Sciences (SPSS) Quality improvement t-test nnIntroduction This text methodically explains how to move from the formulation of the hypothesis to the data collection stage of a research project. Data collection in evidence-based practice (EBP) might be considered the easiest part of the whole research experience. The researcher has already formed the hypothesis, developed the data collection methods and instruments, and determined the subject pool characteristics. Once the EBP researcher has completed data collection, it is time for the researcher to compile and interpret the data so as to explain them in a meaningful context. -

Chapter 4. Data Analysis
4. DATA ANALYSIS Data analysis begins in the monitoring program group of observations selected from the target design phase. Those responsible for monitoring population. In the case of water quality should identify the goals and objectives for monitoring, descriptive statistics of samples are monitoring and the methods to be used for used almost invariably to formulate conclusions or analyzing the collected data. Monitoring statistical inferences regarding populations objectives should be specific statements of (MacDonald et al., 1991; Mendenhall, 1971; measurable results to be achieved within a stated Remington and Schork, 1970; Sokal and Rohlf, time period (Ponce, 1980b). Chapter 2 provides 1981). A point estimate is a single number an overview of commonly encountered monitoring representing the descriptive statistic that is objectives. Once goals and objectives have been computed from the sample or group of clearly established, data analysis approaches can observations (Freund, 1973). For example, the be explored. mean total suspended solids concentration during baseflow is 35 mg/L. Point estimates such as the Typical data analysis procedures usually begin mean (as in this example), median, mode, or with screening and graphical methods, followed geometric mean from a sample describe the central by evaluating statistical assumptions, computing tendency or location of the sample. The standard summary statistics, and comparing groups of data. deviation and interquartile range could likewise be The analyst should take care in addressing the used as point estimates of spread or variability. issues identified in the information expectations report (Section 2.2). By selecting and applying The use of point estimates is warranted in some suitable methods, the data analyst responsible for cases, but in nonpoint source analyses point evaluating the data can prevent the “data estimates of central tendency should be coupled rich)information poor syndrome” (Ward 1996; with an interval estimate because of the large Ward et al., 1986). -

Chapter 5: Spatial Autocorrelation Statistics
Chapter 5: Spatial Autocorrelation Statistics Ned Levine1 Ned Levine & Associates Houston, TX 1 The author would like to thank Dr. David Wong for help with the Getis-Ord >G= and local Getis-Ord statistics. Table of Contents Spatial Autocorrelation 5.1 Spatial Autocorrelation Statistics for Zonal Data 5.3 Indices of Spatial Autocorrelation 5.3 Assigning Point Data to Zones 5.3 Spatial Autocorrelation Statistics for Attribute Data 5.5 Moran=s “I” Statistic 5.5 Adjust for Small Distances 5.6 Testing the Significance of Moran’s “I” 5.7 Example: Testing Houston Burglaries with Moran’s “I” 5.8 Comparing Moran’s “I” for Two Distributions 5.10 Geary=s “C” Statistic 5.10 Adjusted “C” 5.13 Adjust for Small Distances 5.13 Testing the Significance of Geary’s “C” 5.14 Example: Testing Houston Burglaries with Geary’s “C” 5.14 Getis-Ord “G” Statistic 5.16 Testing the Significance of “G” 5.18 Simulating Confidence Intervals for “G” 5.20 Example: Testing Simulated Data with the Getis-Ord AG@ 5.20 Example: Testing Houston Burglaries with the Getis-Ord AG@ 5.25 Use and Limitations of the Getis-Ord AG@ 5.25 Moran Correlogram 5.26 Adjust for Small Distances 5.27 Simulation of Confidence Intervals 5.27 Example: Moran Correlogram of Baltimore County Vehicle Theft and Population 5.27 Uses and Limitations of the Moran Correlogram 5.30 Geary Correlogram 5.34 Adjust for Small Distances 5.34 Geary Correlogram Simulation of Confidence Intervals 5.34 Example: Geary Correlogram of Baltimore County Vehicle Thefts 5.34 Uses and Limitations of the Geary Correlogram 5.35 Table of Contents (continued) Getis-Ord Correlogram 5.35 Getis-Ord Simulation of Confidence Intervals 5.37 Example: Getis-Ord Correlogram of Baltimore County Vehicle Thefts 5.37 Uses and Limitations of the Getis-Ord Correlogram 5.39 Running the Spatial Autocorrelation Routines 5.39 Guidelines for Examining Spatial Autocorrelation 5.39 References 5.42 Attachments 5.44 A. -

STANDARDS and GUIDELINES for STATISTICAL SURVEYS September 2006
OFFICE OF MANAGEMENT AND BUDGET STANDARDS AND GUIDELINES FOR STATISTICAL SURVEYS September 2006 Table of Contents LIST OF STANDARDS FOR STATISTICAL SURVEYS ....................................................... i INTRODUCTION......................................................................................................................... 1 SECTION 1 DEVELOPMENT OF CONCEPTS, METHODS, AND DESIGN .................. 5 Section 1.1 Survey Planning..................................................................................................... 5 Section 1.2 Survey Design........................................................................................................ 7 Section 1.3 Survey Response Rates.......................................................................................... 8 Section 1.4 Pretesting Survey Systems..................................................................................... 9 SECTION 2 COLLECTION OF DATA................................................................................... 9 Section 2.1 Developing Sampling Frames................................................................................ 9 Section 2.2 Required Notifications to Potential Survey Respondents.................................... 10 Section 2.3 Data Collection Methodology.............................................................................. 11 SECTION 3 PROCESSING AND EDITING OF DATA...................................................... 13 Section 3.1 Data Editing ........................................................................................................ -

Treatment Episode Data Set (TEDS) 2001-2011
Treatment Episode Data Set (TEDS) 2001-2011 State Admissions to Substance Abuse Treatment Services DEPARTMENT OF HEALTH AND HUMAN SERVICES Substance Abuse and Mental Health Services Administration Acknowledgments This report was prepared for the Substance Abuse and Mental Health Services Administration (SAMHSA), U.S. Department of Health and Human Services (HHS), by Synectics for Management Decisions, Inc. (Synectics), Arlington, Virginia. Data collection was performed by Mathematica Policy Research (Mathematica), Princeton, New Jersey. Work by Synectics and Mathematica was performed under Task Order HHSS283200700048I/HHSS28342001T, Reference No. 283-07-4803 (Cathie Alderks, Task Order Officer). Public domAin notice All material appearing in this report is in the public domain and may be reproduced or copied without permission from SAMHSA. Citation of the source is appreciated. However, this publication may not be reproduced or distributed for a fee without the specific, written authorization of the Office of Communications, SAMHSA, U.S. Department of Health and Human Services. Recommended citAtion Substance Abuse and Mental Health Services Administration, Center for Behavioral Health Statistics and Quality. Treatment Episode Data Set (TEDS): 2001-2011. State Admissions to Substance Abuse Treatment Services. BHSIS Series S-XX, HHS Publication No. (SMA) XX-XXXX. Rockville, MD: Substance Abuse and Mental Health Services Administration, 2013. electRonic Access And coPies of PublicAtion This publication may be downloaded or ordered at store.samhsa.gov. -

Descriptive Statistics
Statistics: Descriptive Statistics When we are given a large data set, it is necessary to describe the data in some way. The raw data is just too large and meaningless on its own to make sense of. We will sue the following exam scores data throughout: x1, x2, x3, x4, x5, x6, x7, x8, x9, x10 45, 67, 87, 21, 43, 98, 28, 23, 28, 75 Summary Statistics We can use the raw data to calculate summary statistics so that we have some idea what the data looks like and how sprad out it is. Max, Min and Range The maximum value of the dataset and the minimum value of the dataset are very simple measures. The range of the data is difference between the maximum and minimum value. Range = Max Value − Min Value = 98 − 21 = 77 Mean, Median and Mode The mean, median and mode are measures of central tendency of the data (i.e. where is the center of the data). Mean (µ) The mean is sum of all values divided by how many values there are N 1 X 45 + 67 + 87 + 21 + 43 + 98 + 28 + 23 + 28 + 75 xi = = 51.5 N i=1 10 Median The median is the middle data point when the dataset is arranged in order from smallest to largest. If there are two middle values then we take the average of the two values. Using the data above check that the median is: 44 Mode The mode is the value in the dataset that appears most. Using the data above check that the mode is: 28 Standard Deviation The standard deviation (σ) measures how spread out the data is.