Tutorial: an Example of Statistical Data Analysis Using the R Environment for Statistical Computing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Generalized Linear Model for Principal Component Analysis of Binary Data
A Generalized Linear Model for Principal Component Analysis of Binary Data Andrew I. Schein Lawrence K. Saul Lyle H. Ungar Department of Computer and Information Science University of Pennsylvania Moore School Building 200 South 33rd Street Philadelphia, PA 19104-6389 {ais,lsaul,ungar}@cis.upenn.edu Abstract they are not generally appropriate for other data types. Recently, Collins et al.[5] derived generalized criteria We investigate a generalized linear model for for dimensionality reduction by appealing to proper- dimensionality reduction of binary data. The ties of distributions in the exponential family. In their model is related to principal component anal- framework, the conventional PCA of real-valued data ysis (PCA) in the same way that logistic re- emerges naturally from assuming a Gaussian distribu- gression is related to linear regression. Thus tion over a set of observations, while generalized ver- we refer to the model as logistic PCA. In this sions of PCA for binary and nonnegative data emerge paper, we derive an alternating least squares respectively by substituting the Bernoulli and Pois- method to estimate the basis vectors and gen- son distributions for the Gaussian. For binary data, eralized linear coefficients of the logistic PCA the generalized model's relationship to PCA is anal- model. The resulting updates have a simple ogous to the relationship between logistic and linear closed form and are guaranteed at each iter- regression[12]. In particular, the model exploits the ation to improve the model's likelihood. We log-odds as the natural parameter of the Bernoulli dis- evaluate the performance of logistic PCA|as tribution and the logistic function as its canonical link. -

Synthpop: Bespoke Creation of Synthetic Data in R
synthpop: Bespoke Creation of Synthetic Data in R Beata Nowok Gillian M Raab Chris Dibben University of Edinburgh University of Edinburgh University of Edinburgh Abstract In many contexts, confidentiality constraints severely restrict access to unique and valuable microdata. Synthetic data which mimic the original observed data and preserve the relationships between variables but do not contain any disclosive records are one possible solution to this problem. The synthpop package for R, introduced in this paper, provides routines to generate synthetic versions of original data sets. We describe the methodology and its consequences for the data characteristics. We illustrate the package features using a survey data example. Keywords: synthetic data, disclosure control, CART, R, UK Longitudinal Studies. This introduction to the R package synthpop is a slightly amended version of Nowok B, Raab GM, Dibben C (2016). synthpop: Bespoke Creation of Synthetic Data in R. Journal of Statistical Software, 74(11), 1-26. doi:10.18637/jss.v074.i11. URL https://www.jstatsoft. org/article/view/v074i11. 1. Introduction and background 1.1. Synthetic data for disclosure control National statistical agencies and other institutions gather large amounts of information about individuals and organisations. Such data can be used to understand population processes so as to inform policy and planning. The cost of such data can be considerable, both for the collectors and the subjects who provide their data. Because of confidentiality constraints and guarantees issued to data subjects the full access to such data is often restricted to the staff of the collection agencies. Traditionally, data collectors have used anonymisation along with simple perturbation methods such as aggregation, recoding, record-swapping, suppression of sensitive values or adding random noise to prevent the identification of data subjects. -

Bootstrapping Regression Models
Bootstrapping Regression Models Appendix to An R and S-PLUS Companion to Applied Regression John Fox January 2002 1 Basic Ideas Bootstrapping is a general approach to statistical inference based on building a sampling distribution for a statistic by resampling from the data at hand. The term ‘bootstrapping,’ due to Efron (1979), is an allusion to the expression ‘pulling oneself up by one’s bootstraps’ – in this case, using the sample data as a population from which repeated samples are drawn. At first blush, the approach seems circular, but has been shown to be sound. Two S libraries for bootstrapping are associated with extensive treatments of the subject: Efron and Tibshirani’s (1993) bootstrap library, and Davison and Hinkley’s (1997) boot library. Of the two, boot, programmed by A. J. Canty, is somewhat more capable, and will be used for the examples in this appendix. There are several forms of the bootstrap, and, additionally, several other resampling methods that are related to it, such as jackknifing, cross-validation, randomization tests,andpermutation tests. I will stress the nonparametric bootstrap. Suppose that we draw a sample S = {X1,X2, ..., Xn} from a population P = {x1,x2, ..., xN };imagine further, at least for the time being, that N is very much larger than n,andthatS is either a simple random sample or an independent random sample from P;1 I will briefly consider other sampling schemes at the end of the appendix. It will also help initially to think of the elements of the population (and, hence, of the sample) as scalar values, but they could just as easily be vectors (i.e., multivariate). -

A Survey on Data Collection for Machine Learning a Big Data - AI Integration Perspective
1 A Survey on Data Collection for Machine Learning A Big Data - AI Integration Perspective Yuji Roh, Geon Heo, Steven Euijong Whang, Senior Member, IEEE Abstract—Data collection is a major bottleneck in machine learning and an active research topic in multiple communities. There are largely two reasons data collection has recently become a critical issue. First, as machine learning is becoming more widely-used, we are seeing new applications that do not necessarily have enough labeled data. Second, unlike traditional machine learning, deep learning techniques automatically generate features, which saves feature engineering costs, but in return may require larger amounts of labeled data. Interestingly, recent research in data collection comes not only from the machine learning, natural language, and computer vision communities, but also from the data management community due to the importance of handling large amounts of data. In this survey, we perform a comprehensive study of data collection from a data management point of view. Data collection largely consists of data acquisition, data labeling, and improvement of existing data or models. We provide a research landscape of these operations, provide guidelines on which technique to use when, and identify interesting research challenges. The integration of machine learning and data management for data collection is part of a larger trend of Big data and Artificial Intelligence (AI) integration and opens many opportunities for new research. Index Terms—data collection, data acquisition, data labeling, machine learning F 1 INTRODUCTION E are living in exciting times where machine learning expertise. This problem applies to any novel application that W is having a profound influence on a wide range of benefits from machine learning. -

Exploratory and Confirmatory Factor Analysis Course Outline Part 1
Course Outline Exploratory and Confirmatory Factor Analysis 1 Principal components analysis Part 1: Overview, PCA and Biplots FA vs. PCA Least squares fit to a data matrix Biplots Michael Friendly 2 Basic Ideas of Factor Analysis Parsimony– common variance small number of factors. Linear regression on common factors→ Partial linear independence Psychology 6140 Common vs. unique variance 3 The Common Factor Model Factoring methods: Principal factors, Unweighted Least Squares, Maximum λ1 X1 z1 likelihood ξ λ2 Factor rotation X2 z2 4 Confirmatory Factor Analysis Development of CFA models Applications of CFA PCA and Factor Analysis: Overview & Goals Why do Factor Analysis? Part 1: Outline Why do “Factor Analysis”? 1 PCA and Factor Analysis: Overview & Goals Why do Factor Analysis? Data Reduction: Replace a large number of variables with a smaller Two modes of Factor Analysis number which reflect most of the original data [PCA rather than FA] Brief history of Factor Analysis Example: In a study of the reactions of cancer patients to radiotherapy, measurements were made on 10 different reaction variables. Because it 2 Principal components analysis was difficult to interpret all 10 variables together, PCA was used to find Artificial PCA example simpler measure(s) of patient response to treatment that contained most of the information in data. 3 PCA: details Test and Scale Construction: Develop tests and scales which are “pure” 4 PCA: Example measures of some construct. Example: In developing a test of English as a Second Language, 5 Biplots investigators calculate correlations among the item scores, and use FA to Low-D views based on PCA construct subscales. -

A Resampling Test for Principal Component Analysis of Genotype-By-Environment Interaction
A RESAMPLING TEST FOR PRINCIPAL COMPONENT ANALYSIS OF GENOTYPE-BY-ENVIRONMENT INTERACTION JOHANNES FORKMAN Abstract. In crop science, genotype-by-environment interaction is of- ten explored using the \genotype main effects and genotype-by-environ- ment interaction effects” (GGE) model. Using this model, a singular value decomposition is performed on the matrix of residuals from a fit of a linear model with main effects of environments. Provided that errors are independent, normally distributed and homoscedastic, the signifi- cance of the multiplicative terms of the GGE model can be tested using resampling methods. The GGE method is closely related to principal component analysis (PCA). The present paper describes i) the GGE model, ii) the simple parametric bootstrap method for testing multi- plicative genotype-by-environment interaction terms, and iii) how this resampling method can also be used for testing principal components in PCA. 1. Introduction Forkman and Piepho (2014) proposed a resampling method for testing in- teraction terms in models for analysis of genotype-by-environment data. The \genotype main effects and genotype-by-environment interaction ef- fects"(GGE) analysis (Yan et al. 2000; Yan and Kang, 2002) is closely related to principal component analysis (PCA). For this reason, the method proposed by Forkman and Piepho (2014), which is called the \simple para- metric bootstrap method", can be used for testing principal components in PCA as well. The proposed resampling method is parametric in the sense that it assumes homoscedastic and normally distributed observations. The method is \simple", because it only involves repeated sampling of standard normal distributed values. Specifically, no parameters need to be estimated. -

GGL Biplot Analysis of Durum Wheat (Triticum Turgidum Spp
756 Bulgarian Journal of Agricultural Science, 19 (No 4) 2013, 756-765 Agricultural Academy GGL BIPLOT ANALYSIS OF DURUM WHEAT (TRITICUM TURGIDUM SPP. DURUM) YIELD IN MULTI-ENVIRONMENT TRIALS N. SABAGHNIA2*, R. KARIMIZADEH1 and M. MOHAMMADI1 1 Department of Agronomy and Plant Breeding, Faculty of Agriculture, University of Maragheh, Maragheh, Iran 2 Dryland Agricultural Research Institute (DARI), Gachsaran, Iran Abstract SABAGHNIA, N., R. KARIMIZADEH and M. MOHAMMADI, 2013. GGL biplot analysis of durum wheat (Triticum turgidum spp. durum) yield in multi-environment trials. Bulg. J. Agric. Sci., 19: 756-765 Durum wheat (Triticum turgidum spp. durum) breeders have to determine the new genotypes responsive to the environ- mental changes for grain yield. Matching durum wheat genotype selection with its production environment is challenged by the occurrence of significant genotype by environment (GE) interaction in multi-environment trials (MET). This investigation was conducted to evaluate 20 durum wheat genotypes for their stability grown in five different locations across three years using randomized completely block design with 4 replications. According to combined analysis of variance, the main effects of genotypes, locations and years, were significant as well as the interactions effects. The first two principal components of the site regression model accounted for 60.3 % of the total variation. Polygon view of genotype plus genotype by location (GGL) biplot indicated that there were three winning genotypes in three mega-environments for durum wheat in rain-fed conditions. Genotype G14 was the most favorable genotype for location Gachsaran and the most favorable genotype of mega-environment Kouhdasht and Ilam was G12 while G10 was the most favorable genotypes for mega-environment Gonbad and Moghan. -

A First Step Into the Bootstrap World
INSTITUTE FOR DEFENSE ANALYSES A First Step into the Bootstrap World Matthew Avery May 2016 Approved for public release. IDA Document NS D-5816 Log: H 16-000624 INSTITUTE FOR DEFENSE ANALYSES 4850 Mark Center Drive Alexandria, Virginia 22311-1882 The Institute for Defense Analyses is a non-profit corporation that operates three federally funded research and development centers to provide objective analyses of national security issues, particularly those requiring scientific and technical expertise, and conduct related research on other national challenges. About This Publication This briefing motivates bootstrapping through an intuitive explanation of basic statistical inference, discusses the right way to resample for bootstrapping, and uses examples from operational testing where the bootstrap approach can be applied. Bootstrapping is a powerful and flexible tool when applied appropriately. By treating the observed sample as if it were the population, the sampling distribution of statistics of interest can be generated with precision equal to Monte Carlo error. This approach is nonparametric and requires only acceptance of the observed sample as an estimate for the population. Careful resampling is used to generate the bootstrap distribution. Applications include quantifying uncertainty for availability data, quantifying uncertainty for complex distributions, and using the bootstrap for inference, such as the two-sample t-test. Copyright Notice © 2016 Institute for Defense Analyses 4850 Mark Center Drive, Alexandria, Virginia 22311-1882 -
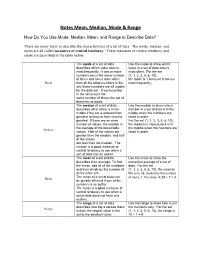
Notes Mean, Median, Mode & Range
Notes Mean, Median, Mode & Range How Do You Use Mode, Median, Mean, and Range to Describe Data? There are many ways to describe the characteristics of a set of data. The mode, median, and mean are all called measures of central tendency. These measures of central tendency and range are described in the table below. The mode of a set of data Use the mode to show which describes which value occurs value in a set of data occurs most frequently. If two or more most often. For the set numbers occur the same number {1, 1, 2, 3, 5, 6, 10}, of times and occur more often the mode is 1 because it occurs Mode than all the other numbers in the most frequently. set, those numbers are all modes for the data set. If each number in the set occurs the same number of times, the set of data has no mode. The median of a set of data Use the median to show which describes what value is in the number in a set of data is in the middle if the set is ordered from middle when the numbers are greatest to least or from least to listed in order. greatest. If there are an even For the set {1, 1, 2, 3, 5, 6, 10}, number of values, the median is the median is 3 because it is in the average of the two middle the middle when the numbers are Median values. Half of the values are listed in order. greater than the median, and half of the values are less than the median. -

A Biplot-Based PCA Approach to Study the Relations Between Indoor and Outdoor Air Pollutants Using Case Study Buildings
buildings Article A Biplot-Based PCA Approach to Study the Relations between Indoor and Outdoor Air Pollutants Using Case Study Buildings He Zhang * and Ravi Srinivasan UrbSys (Urban Building Energy, Sensing, Controls, Big Data Analysis, and Visualization) Laboratory, M.E. Rinker, Sr. School of Construction Management, University of Florida, Gainesville, FL 32603, USA; sravi@ufl.edu * Correspondence: rupta00@ufl.edu Abstract: The 24 h and 14-day relationship between indoor and outdoor PM2.5, PM10, NO2, relative humidity, and temperature were assessed for an elementary school (site 1), a laboratory (site 2), and a residential unit (site 3) in Gainesville city, Florida. The primary aim of this study was to introduce a biplot-based PCA approach to visualize and validate the correlation among indoor and outdoor air quality data. The Spearman coefficients showed a stronger correlation among these target environmental measurements on site 1 and site 2, while it showed a weaker correlation on site 3. The biplot-based PCA regression performed higher dependency for site 1 and site 2 (p < 0.001) when compared to the correlation values and showed a lower dependency for site 3. The results displayed a mismatch between the biplot-based PCA and correlation analysis for site 3. The method utilized in this paper can be implemented in studies and analyzes high volumes of multiple building environmental measurements along with optimized visualization. Keywords: air pollution; indoor air quality; principal component analysis; biplot Citation: Zhang, H.; Srinivasan, R. A Biplot-Based PCA Approach to Study the Relations between Indoor and 1. Introduction Outdoor Air Pollutants Using Case The 2020 Global Health Observatory (GHO) statistics show that indoor and outdoor Buildings 2021 11 Study Buildings. -

Common Data Set 2020-2021 University of Pennsylvania
Common Data Set 2020-2021 University of Pennsylvania Table of Contents A General Information page 3 B Enrollment and Persistence 5 C First Time, First Year Admission 8 D Transfer Admission 14 E Academic Offerings and Policies 16 F Student Life 18 G Annual Expenses 20 H Financial Aid 22 I Instructional Faculty and Class Size 28 J Disciplinary Areas of Degrees Conferred 30 Common Data Set Definitions 32 Common Data Set 2020-21 1 25 Jun 2021 Common Data Set 2020-21 2 25 Jun 2021 A. General Information return to Table of Contents A1 Address Information A1 University of Pennsylvania A1 Mailing Address: 1 College Hall, Room 100 A1 City/State/Zip/Country: Philadelphia, PA 19104-6228 A1 Main Phone Number: 215-898-5000 A1 Home Page Address www.upenn.edu A1 Admissions Phone Number: 215-898-7507 A1 Admissions Office Mailing Address: 1 College Hall A1 City/State/Zip/Country: Philadelphia, PA 19104 A1 Admissions Fax Number: 215-898-9670 A1 Admissions E-mail Address: [email protected] A1 Online application www.admissions.upenn.edu A2 Source of institutional control (Check only one): A2 Public A2 Private (nonprofit) x A2 Proprietary A3 Classify your undergraduate institution: A3 Coeducational college A3 Men's college A3 Women's college A4 Academic year calendar: A4 Semester x A4 Quarter A4 Trimester A4 4-1-4 A4 Continuous A5 Degrees offered by your institution: A5 Certificate x A5 Diploma A5 Associate x A5 Transfer Associate A5 Terminal Associate x A5 Bachelor's x A5 Postbachelor's certificate x A5 Master's x A5 Post-master's certificate x A5 Doctoral degree - research/scholarship x A5 Doctoral degree - professional practice x A5 Doctoral degree - other x A5 Doctoral degree -- other Common Data Set 2020-21 3 25 Jun 2021 Common Data Set 2020-21 4 25 Jun 2021 B. -

Logistic Biplot by Conjugate Gradient Algorithms and Iterated SVD
mathematics Article Logistic Biplot by Conjugate Gradient Algorithms and Iterated SVD Jose Giovany Babativa-Márquez 1,2,* and José Luis Vicente-Villardón 1 1 Department of Statistics, University of Salamanca, 37008 Salamanca, Spain; [email protected] 2 Facultad de Ciencias de la Salud y del Deporte, Fundación Universitaria del Área Andina, Bogotá 1321, Colombia * Correspondence: [email protected] Abstract: Multivariate binary data are increasingly frequent in practice. Although some adaptations of principal component analysis are used to reduce dimensionality for this kind of data, none of them provide a simultaneous representation of rows and columns (biplot). Recently, a technique named logistic biplot (LB) has been developed to represent the rows and columns of a binary data matrix simultaneously, even though the algorithm used to fit the parameters is too computationally demanding to be useful in the presence of sparsity or when the matrix is large. We propose the fitting of an LB model using nonlinear conjugate gradient (CG) or majorization–minimization (MM) algo- rithms, and a cross-validation procedure is introduced to select the hyperparameter that represents the number of dimensions in the model. A Monte Carlo study that considers scenarios with several sparsity levels and different dimensions of the binary data set shows that the procedure based on cross-validation is successful in the selection of the model for all algorithms studied. The comparison of the running times shows that the CG algorithm is more efficient in the presence of sparsity and when the matrix is not very large, while the performance of the MM algorithm is better when the binary matrix is balanced or large.