Danmap 2006.Pmd
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Oral Presentations September 23Rd - Rooms 1,2 and 3
Oral Presentations September 23rd - Rooms 1,2 and 3 Presentation Date Abstract Authors Presenter´s name - Theme Title Code indicated by the author 18498 Thomas Smits; Femke Gresnigt; Thomas Smits Clinical Toxicology/drugs of PERFORMANCE OF AN IMMUNOASSAY Eric Franssen; Milly Attema-de abuse METHOD FOR GAMMA-HYDROXYBUTYRIC Jonge ACID (GHB) IN PATIENTS PRESENTED AT THE EMERGENCY DEPARTMENT, A PROSPECTIVE STUDY 18499 Thomas Smits; Femke Gresnigt; Thomas Smits Clinical Toxicology/drugs of DO WE NEED POINT-OF-CARE TESTING OF Milly Attema-de Jonge; Eric abuse GAMMA-HYDROXYBUTYRIC ACID (GHB) AT Fransse THE EMERGENCY DEPARTMENT? September 23 18730 Lilian H.J. Richter; Julia Menges; Lea Wagmann Clinical Toxicology/drugs of NEW PSYCHOACTIVE SUBSTANCES: Lea Wagmann; Simon D. Brandt; abuse METABOLIC FATE, ISOZYME-MAPPING, 13:30 - 14:45 Folker Westphal; Veit Flockerzi; AND PLASMA PROTEIN BINDING OF 5-APB- ROOM 1 Markus R. Meyer NBOME, 2C-B-FLY-NB2ETO5CL, AND 2C-B- FLY-NBOME 18985 Annelies Cannaert; Marie Annelies Cannaert Clinical Toxicology/drugs of HIDE AND SEEK: OVERCOMING THE Deventer; Melissa Fogarty; abuse MASKING EFFECT OF OPIOID Amanda L.A. Mohr; Christophe P. ANTAGONISTS IN ACTIVITY-BASED Stove SCREENING TESTS 18740 Souleiman El Balkhi ; Roland Souleiman El Balkhi Clinical Toxicology/drugs of METABOLIC INTERACTIONS BETWEEN Lawson; Franck Saint-Marcoux abuse OXYCODONE, BENZODIAZEPINES OR DESIGNER BENZODIAZEPINES PLAY AN IMPORTANT ROLE IN OXYCODONE INTOXICATIONS 19050 Brenda de Winter F de Velde; MN Brenda de Winter Anti-infective drugs POPULATION -
Therapeutic Alternatives for Drug-Resistant Cabapenemase- Producing Enterobacteria
THERAPEUTIC ALTERNATIVES FOR MULTIDRUG-RESISTANT AND EXTREMELY All isolates resistant to carbapenems were evaluated Fig. 1: Susceptibility (%) of Carbapenemase-producing MDRE Fig. 3: Susceptibility (%) of VIM-producing MDRE to Alternative DRUG-RESISTANT CABAPENEMASE- for the presence of genes encoding carbapenemases to Alternative Antibiotics Antibiotics PRODUCING ENTEROBACTERIA OXA-48-like, KPC, GES, NDM, VIM, IMP and GIM. M. Almagro*, A. Kramer, B. Gross, S. Suerbaum, S. Schubert. Max Von Pettenkofer Institut, Faculty of Medicine. LMU Munich, München, Germany. RESULTS BACKROUND 44 isolates of CPE were collected: OXA-48 (n=29), VIM (n=9), NDM-1 (n=5), KPC (n=1) and GES (n=1). The increasing emergence and dissemination of carbapenem-resistant gram-negative bacilli has From the 44 CPE isolates, 26 isolates were identified reduced significantly the options for sufficient as Klebsiella pneumoniae (68% of the OXA-48 CPE), 8 Fig. 2: Susceptibility (%) of OXA-48-producing MDRE to Fig. 4: Susceptibility (%) of NDM-producing MDRE to Alternative antibiotic therapy. The genes encoding most of these as Escherichia coli, 6 as Enterobacter cloacae, 2 as Alternative Antibiotics Antibiotics carbapenemases reside on plasmids or transposons Citrobacter freundii,1 as Providencia stuartii and 1 as carrying additional resistance genes which confer Morganella morganii. multidrug resistance to the isolates. 31 isolates (70%) were causing an infection, including urinary tract infection (20%), respiratory tract MATERIALS AND METHODS infection (18%), abdominal infection (18%), bacteraemia (9%) and skin and soft tissue infection In the present study, we tested the in vitro activity of (5%). 13 isolates were believed to be colonizers. antimicrobial agents against a well-characterized c o l l e c t i o n o f c a r b a p e n e m a s e - p r o d u c i n g Isolates were classified as 32 MDRE and 13 XDRE. -

National Antibiotic Consumption for Human Use in Sierra Leone (2017–2019): a Cross-Sectional Study
Tropical Medicine and Infectious Disease Article National Antibiotic Consumption for Human Use in Sierra Leone (2017–2019): A Cross-Sectional Study Joseph Sam Kanu 1,2,* , Mohammed Khogali 3, Katrina Hann 4 , Wenjing Tao 5, Shuwary Barlatt 6,7, James Komeh 6, Joy Johnson 6, Mohamed Sesay 6, Mohamed Alex Vandi 8, Hannock Tweya 9, Collins Timire 10, Onome Thomas Abiri 6,11 , Fawzi Thomas 6, Ahmed Sankoh-Hughes 12, Bailah Molleh 4, Anna Maruta 13 and Anthony D. Harries 10,14 1 National Disease Surveillance Programme, Sierra Leone National Public Health Emergency Operations Centre, Ministry of Health and Sanitation, Cockerill, Wilkinson Road, Freetown, Sierra Leone 2 Department of Community Health, Faculty of Clinical Sciences, College of Medicine and Allied Health Sciences, University of Sierra Leone, Freetown, Sierra Leone 3 Special Programme for Research and Training in Tropical Diseases (TDR), World Health Organization, 1211 Geneva, Switzerland; [email protected] 4 Sustainable Health Systems, Freetown, Sierra Leone; [email protected] (K.H.); [email protected] (B.M.) 5 Unit for Antibiotics and Infection Control, Public Health Agency of Sweden, Folkhalsomyndigheten, SE-171 82 Stockholm, Sweden; [email protected] 6 Pharmacy Board of Sierra Leone, Central Medical Stores, New England Ville, Freetown, Sierra Leone; [email protected] (S.B.); [email protected] (J.K.); [email protected] (J.J.); [email protected] (M.S.); [email protected] (O.T.A.); [email protected] (F.T.) Citation: Kanu, J.S.; Khogali, M.; 7 Department of Pharmaceutics and Clinical Pharmacy & Therapeutics, Faculty of Pharmaceutical Sciences, Hann, K.; Tao, W.; Barlatt, S.; Komeh, College of Medicine and Allied Health Sciences, University of Sierra Leone, Freetown 0000, Sierra Leone 8 J.; Johnson, J.; Sesay, M.; Vandi, M.A.; Directorate of Health Security & Emergencies, Ministry of Health and Sanitation, Sierra Leone National Tweya, H.; et al. -
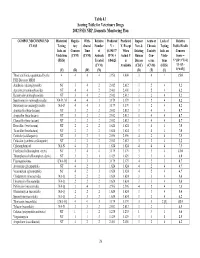
2002 FSIS National Residue Program, Section 4
Table 4.1 Scoring Table for Veterinary Drugs 2002 FSIS NRP, Domestic Monitoring Plan COMPOUND/COMPOUND Historical Regula- With- Relative Predicted Predicted Impact Acute or Lack of Relative CLASS Testing tory drawal Number V = V, Except New & Chronic Testing Public Health Info. on Concern Time of (0.19437* When Existing Toxicity Info. on Concern Violations (CVM) (CVM) Animals R*N) + Actual V Human Con- Viola- Score = (FSIS) Treated 0.84625 is Disease cerns tions V*[(D+3*T)/4] (CVM) Available (CDC) (CVM) (FSIS) *{1+[(L- (V) (R) (W) (N) (D) (T) (L) 1)*0.05]} Those antibiotics quantitated by the 4 4 4 4 3.956 4.000 3 4 1 15.0 FSIS Bioassay MRM Amikacin (aminoglycoside) NT 3 4 2 2.012 2.012 3 2 4 5.2 Apramycin (aminoglycoside) NT 4 4 2 2.401 2.401 3 2 4 6.2 Kanamycin (aminoglycoside) NT 3 4 2 2.012 2.012 3 2 4 5.2 Spectinomycin (aminoglycoside) NA-D, M 4 4 3 3.179 3.179 3 2 4 8.2 Streptomycin (aminoglycoside) NA-D 4 4 3 3.179 3.179 3 2 4 8.2 Amoxicillin (beta-lactam) NT 3 2 2 2.012 2.012 3 4 4 8.7 Ampicillin (beta-lactam) NT 3 2 2 2.012 2.012 3 4 4 8.7 Cloxacillin (beta-lactam) NT 3 2 2 2.012 2.012 3 4 4 8.7 Hetacillin (beta-lactam) NT 2 2 2 1.624 1.624 3 4 4 7.0 Ticarcillin (beta-lactam) NT 2 2 2 1.624 1.624 3 4 4 7.0 Ceftiofur (cefalosporin) NT 3 2 3 2.596 2.596 4 2 4 7.5 Cefazolin (synthetic cefalosporin) NT 3 2 2 2.012 2.012 3 2 4 5.2 Chloramphenicol NA-N 4 2 1 1.624 1.624 4 4 4 7.5 Florfenicol (chloramphen. -

NARMS – EB 2000 Veterinary Isolates Fig. 18. Resistance Among Salmonella Serotypes for Isolates from Cattle*
NARMS – EB 2000 Veterinary Isolates Fig. 18. Resistance Among Salmonella Serotypes for Isolates from Cattle* S. typhimurium (n=332)** S. montevideo (n=330) Amikacin 0.00% Amikacin 0.00% Amox-Clav 14.46% Amox-Clav 0.61% Ampicillin 66.87% Ampicillin 0.61% Apramycin 1.20% Apramycin 0.00% Cefoxitin 10.54% Cefoxitin 0.61% Ceftiofur 12.65% Ceftiofur 0.61% Ceftriaxone 0.00% Ceftriaxone 0.00% Cephalothin 13.25% Cephalothin 0.61% Chloramphenicol 39.46% Chloramphenicol 0.91% Ciprofloxacin 0.00% Ciprofloxacin 0.00% Gentamicin 3.31% Gentamicin 0.30% Kanamycin 38.86% Kanamycin 0.00% Nalidixic Acid 0.00% Nalidixic Acid 0.00% Streptomycin 66.57% Streptomycin 1.52% Sulfamethoxazole 66.87% Sulfamethoxazole 1.21% Tetracycline 66.57% Tetracycline 2.12% Trimeth-Sulfa 5.42% Trimeth-Sulfa 0.30% 0% 10% 20% 30% 40% 50% 60% 70% 80% 0% 1% 1% 2% 2% 3% **including copenhagen S. anatum (n=292) S. newport (n=185) Amikacin 0.00% Amikacin 0.00% Amox-Clav 1.71% Amox-Clav 80.00% Ampicillin 2.40% Ampicillin 80.54% Apramycin 0.00% Apramycin 0.00% Cefoxitin 1.37% Cefoxitin 77.84% Ceftiofur 1.37% Ceftiofur 80.00% Ceftriaxone 0.00% Ceftriaxone 2.16% Cephalothin 2.05% Cephalothin 77.84% Chloramphenicol 1.71% Chloramphenicol 81.62% Ciprofloxacin 0.00% Ciprofloxacin 0.00% Gentamicin 0.68% Gentamicin 7.57% Kanamycin 1.71% Kanamycin 7.57% Nalidixic Acid 0.34% Nalidixic Acid 0.00% Streptomycin 2.05% Streptomycin 82.70% Sulfamethoxazole 2.05% Sulfamethoxazole 74.05% Tetracycline 35.96% Tetracycline 83.24% Trimeth-Sulfa 0.34% Trimeth-Sulfa 25.41% 0% 5% 10% 15% 20% 25% 30% 35% 40% 0% 10% 20% 30% 40% 50% 60% 70% 80% 90% *all sources NARMS – EB 2000 Veterinary Isolates Fig. -

Infant Antibiotic Exposure Search EMBASE 1. Exp Antibiotic Agent/ 2
Infant Antibiotic Exposure Search EMBASE 1. exp antibiotic agent/ 2. (Acedapsone or Alamethicin or Amdinocillin or Amdinocillin Pivoxil or Amikacin or Aminosalicylic Acid or Amoxicillin or Amoxicillin-Potassium Clavulanate Combination or Amphotericin B or Ampicillin or Anisomycin or Antimycin A or Arsphenamine or Aurodox or Azithromycin or Azlocillin or Aztreonam or Bacitracin or Bacteriocins or Bambermycins or beta-Lactams or Bongkrekic Acid or Brefeldin A or Butirosin Sulfate or Calcimycin or Candicidin or Capreomycin or Carbenicillin or Carfecillin or Cefaclor or Cefadroxil or Cefamandole or Cefatrizine or Cefazolin or Cefixime or Cefmenoxime or Cefmetazole or Cefonicid or Cefoperazone or Cefotaxime or Cefotetan or Cefotiam or Cefoxitin or Cefsulodin or Ceftazidime or Ceftizoxime or Ceftriaxone or Cefuroxime or Cephacetrile or Cephalexin or Cephaloglycin or Cephaloridine or Cephalosporins or Cephalothin or Cephamycins or Cephapirin or Cephradine or Chloramphenicol or Chlortetracycline or Ciprofloxacin or Citrinin or Clarithromycin or Clavulanic Acid or Clavulanic Acids or clindamycin or Clofazimine or Cloxacillin or Colistin or Cyclacillin or Cycloserine or Dactinomycin or Dapsone or Daptomycin or Demeclocycline or Diarylquinolines or Dibekacin or Dicloxacillin or Dihydrostreptomycin Sulfate or Diketopiperazines or Distamycins or Doxycycline or Echinomycin or Edeine or Enoxacin or Enviomycin or Erythromycin or Erythromycin Estolate or Erythromycin Ethylsuccinate or Ethambutol or Ethionamide or Filipin or Floxacillin or Fluoroquinolones -

A Sensitive Method for Direct Analysis of Impurities in Apramycin and Other Aminoglycoside Antibiotics Using Charged Aerosol
- SO4 A Sensitive Method for Direct Analysis of Impurities in Apramycin and Other Aminoglycoside Antibiotics Using Charged Aerosol Detection Zhen Long1, Qi Zhang2, Yan Jin1, Lina Liang1, Bruce Bailey2, Ian Acworth2, Deepali Mohindra3 1 2 3 Thermo Fisher Scientific, Shanghai, China, Thermo Fisher Scientific, Chelmsford, MA, USA and Thermo Fisher Scientific, Sunnyvale, CA, USA Comparison of CAD and ELSD Detection Analysis of Other Aminoglycoside Antibiotics Overview Results Purpose: To develop a sensitive non-derivatization method for impurity assessment of Sample pre-treatment with SPE This method has also been applied to impurity analysis of an additional eleven apramycin sulfate and other aminoglycoside antibiotics. Sample pre-treatment with SPE SPE Column: Dionex OnGuard II A CAD and ELSD are both nebulization-based universal detection technologies. aminoglycoside antibiotics, including neomycin, gentamicin, kanamycin, streptomycin, Comparison between CAD and ELSD under the same chromatographic conditions tobramycin, amikacin, etimicin, netilmicin, sisomicin, ribostamycin and paromomycin. Methods: A 30min gradient method using hydrophilic interaction liquid chromatography Sample solvent: 80% 5mM ammonium formate, 20% acetonitrile Sulfate is a major interference for apramycin impurity assessment with a HILIC method. demonstrated that CAD is much more sensitive than ELSD. As shown in the Figure 5 shows impurity analysis of kenamycin, etimicin, ribostamycin and paromomycin with charged aerosol detection (HILIC-CAD) was developed for direct analysis of Sample: 220.6 mg/mL in 2 mL sample solvent Without sample cleanup, some early eluting impurities were found to be masked under chromatograms in Figure 4 and data summarized in Table 1, 16 impurities (S/N >3) were using the HILIC-CAD method. -

Antimicrobial Resistance Benchmark 2020 Antimicrobial Resistance Benchmark 2020
First independent framework for assessing pharmaceutical company action Antimicrobial Resistance Benchmark 2020 Antimicrobial Resistance Benchmark 2020 ACKNOWLEDGEMENTS The Access to Medicine Foundation would like to thank the following people and organisations for their contributions to this report.1 FUNDERS The Antimicrobial Resistance Benchmark research programme is made possible with financial support from UK AID and the Dutch Ministry of Health, Welfare and Sport. Expert Review Committee Research Team Reviewers Hans Hogerzeil - Chair Gabrielle Breugelmans Christine Årdal Gregory Frank Fatema Rafiqi Karen Gallant Nina Grundmann Adrián Alonso Ruiz Hans Hogerzeil Magdalena Kettis Ruth Baron Hitesh Hurkchand Joakim Larsson Dulce Calçada Joakim Larsson Marc Mendelson Moska Hellamand Marc Mendelson Margareth Ndomondo-Sigonda Kevin Outterson Katarina Nedog Sarah Paulin (Observer) Editorial Team Andrew Singer Anna Massey Deirdre Cogan ACCESS TO MEDICINE FOUNDATION Rachel Jones The Access to Medicine Foundation is an independent Emma Ross non-profit organisation based in the Netherlands. It aims to advance access to medicine in low- and middle-income Additional contributors countries by stimulating and guiding the pharmaceutical Thomas Collin-Lefebvre industry to play a greater role in improving access to Alex Kong medicine. Nestor Papanikolaou Address Contact Naritaweg 227-A For more information about this publication, please contact 1043 CB, Amsterdam Jayasree K. Iyer, Executive Director The Netherlands [email protected] +31 (0) 20 215 35 35 www.amrbenchmark.org 1 This acknowledgement is not intended to imply that the individuals and institutions referred to above endorse About the cover: Young woman from the Antimicrobial Resistance Benchmark methodology, Brazil, where 40%-60% of infections are analyses or results. -

High Level Aminoglycoside Resistance in Enterococcus, Pediococcus and Lactobacillus Species from Farm Animals and Commercial Meat Products
Ann Microbiol (2016) 66:101–110 DOI 10.1007/s13213-015-1086-1 ORIGINAL ARTICLE High level aminoglycoside resistance in Enterococcus, Pediococcus and Lactobacillus species from farm animals and commercial meat products George Jaimee1 & Prakash M. Halami1 Received: 10 September 2014 /Accepted: 8 April 2015 /Published online: 2 May 2015 # Springer-Verlag Berlin Heidelberg and the University of Milan 2015 Abstract Inappropriate use of aminoglycosides in animal I40a suggesting its involvement in antibiotic resistant gene husbandry has led to the selection and emergence of high- transfer. Besides, strains of L. plantarum, a species used as level aminoglycoside resistance (HLAR) in lactic acid bacte- probiotic, isolated in this study showed the occurrence of ria (LAB). The objective of this study was to assess the pres- aph(3′)IIIa as well as aac (6′)Ie-aph(2″)Ia genes that could ence of aminoglycoside resistant LAB in farm animals and be of concern in human health. The findings of the study meat products. Gentamicin resistant LAB (n=138) were se- highlight the spread and emergence of multi-resistance genes lectively isolated from 50 different meat and farm animal for aminoglycoside antibiotics among beneficial LAB. sources. These native isolates of LAB were subsequently characterized for their minimum inhibitory concentration to Keywords Lactic acid bacteria . Minimum inhibitory seven different aminoglycoside antibiotics. HLAR to genta- concentration . Aminoglycoside . Random amplified micin, kanamycin and streptomycin was found to be 38 %, polymorphic DNA . Integrase 45 % and 15 %, respectively. Selected cultures of LAB were identified by random amplified polymorphic DNA (RAPD)-PCR and 16S rDNA gene sequencing. Subsequent Introduction detection for the presence of nine aminoglycoside modifying genes [aac(6′)Ie-aph(2″)Ia, aph(3′)IIIa, aad6, ant(6)Ia, The spread of resistance to antibiotics among bacteria is ant(9)Ia, ant(9)Ib, aph(2″)Ib, aph(2″)Ic and aph(2″)Id] was alarming. -

Identification of Macrolide Antibiotic-Binding Human P8 Protein
J. Antibiot. 61(5): 291–296, 2008 THE JOURNAL OF ORIGINAL ARTICLE ANTIBIOTICS Identification of Macrolide Antibiotic-binding Human_p8 Protein Tetsuro Morimura, Mio Hashiba, Hiroshi Kameda, Mihoko Takami, Hirokazu Takahama, Masahiko Ohshige, Fumio Sugawara Received: December 10, 2007 / Accepted: May 1, 2008 © Japan Antibiotics Research Association Abstract Clarithromycin is a macrolide antibiotic that is widely used in clinical medicine. Macrolide antibiotics Introduction such as clarithromycin specifically bind to the 50S subunit of the bacterial ribosome thereby interfering with protein Launched in 1990 by Abbott Laboratories, clarithromycin biosynthesis. A selected peptide sequence from our former A (CLA: synonym of 6-O-methylerythromycin, 1) [1] is a study, composed of 19 amino acids, which was isolated macrolide antibiotic that is commonly used to treat a from a phage display library because of its ability to bind variety of human bacterial infections. Macrolide clarithromycin, displayed significant similarity to a portion antibiotics, represented by erythromycin A (ERY, 2) and its of the human_p8 protein. The recombinant p8 protein structural homologues, specifically bind to the bacterial binds to biotinylated-clarithromycin immobilized on a 50S ribosomal subunit resulting in the inhibition of streptavidin-coated sensor chip and the dissociation bacterial protein biosynthesis [2]. CLA (1) is also constant was determined. The binding of recombinant p8 commonly used to target gastric Helicobacter pylori, which protein to double-stranded DNA was inhibited by is implicated in both gastric ulcers and cancer [3]. biotinylated-clarithromycin, clarithromycin, erythromycin Numerous studies to develop novel pharmaceutical and azithromycin in gel mobility shift assay. applications for macrolide antibiotics, including CLA (1), Dechlorogriseofulvin, obtained from a natural product ERY (2) and azithromycin (AZM, 3), have been published screening, also inhibited human p8 protein binding to [4]. -

Antibiotic Resistance and Typing of the Methicillin-Resistant Staphylococcus Aureus Clones in Kuwait Hospitals, 2016–2017 Samar S
Boswihi et al. BMC Microbiology (2020) 20:314 https://doi.org/10.1186/s12866-020-02009-w RESEARCH ARTICLE Open Access Antibiotic resistance and typing of the methicillin-resistant Staphylococcus aureus clones in Kuwait hospitals, 2016–2017 Samar S. Boswihi, Edet E. Udo* and Wadha AlFouzan Abstract Background: Methicillin-resistant Staphylococcus aureus (MRSA) belong to diverse genetic backgrounds that differ in antibiotic resistance. Knowledge of the local clonal composition of MRSA strains is important for patients’ management and for designing effective control and eradication methods. The aim of this study was to compare the antibiotic resistance patterns and genotypic characteristics of MRSA isolates obtained in public hospitals in Kuwait in 2016 and 2017 for changes in their resistance patterns and clonal composition. Methods: A total of 4726 MRSA isolates obtained in 2016–2017 from clinical specimens in Kuwait public hospitals were characterized using antibiogram, SCCmec typing, spa typing and DNA microarray. Results: The isolates expressed resistance to fusidic acid (52.9%), kanamycin (41.6%), gentamicin (32.5%) and erythromycin (36.2%). The prevalence of high-level mupirocin resistance decreased from 3.7% in 2016 to 2.4% in 2017, while the proportion of resistance to other antibiotics remained relatively stable. A total of 382 spa types were detected with eight spa types, t688 (N = 547), t304 (N = 428), t860 (N = 394), t127 (N = 306), t044 (N = 230), t311 (N = 243), t223 (N = 184) and t002 (N = 181) constituting 53.1% of the MRSA isolates in 2016–2017. Of the 3004 MRSA isolates obtained in 2016 (N = 1327) and 2017 (N = 1677) selected for DNA microarray analysis, 26 clonal complexes (CCs) were identified. -

Aminoglycosides for Intra-Abdominal Infection: Equal to the Challenge? John A
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by VCU Scholars Compass Virginia Commonwealth University VCU Scholars Compass Publications from the Office of the Dean Office of the Dean 2004 Aminoglycosides for Intra-Abdominal Infection: Equal to the Challenge? John A. Bailey Saint Louis University Katherine S. Virgo Saint Louis University Joseph T. DiPiro Virginia Commonwealth University, University of Georgia, [email protected] See next page for additional authors Follow this and additional works at: http://scholarscompass.vcu.edu/pharmacy_dean_pubs Part of the Pharmacy and Pharmaceutical Sciences Commons This is a copy of an article published in Surgical Infections © 2004 copyright Mary Ann Liebert, Inc.; Surgical Infections is available online at: http://online.liebertpub.com. Downloaded from http://scholarscompass.vcu.edu/pharmacy_dean_pubs/13 This Article is brought to you for free and open access by the Office of the Dean at VCU Scholars Compass. It has been accepted for inclusion in Publications from the Office of the Dean by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Authors John A. Bailey, Katherine S. Virgo, Joseph T. DiPiro, Avery B. Nathens, Robert G. Sawyer, and John E. Mazuski This article is available at VCU Scholars Compass: http://scholarscompass.vcu.edu/pharmacy_dean_pubs/13 SURGICAL INFECTIONS Volume 3, Number 4, 2002 © Mary Ann Liebert, Inc. Aminoglycosides for Intra-Abdominal Infection: Equal to the Challenge? JEFFREY A. BAILEY, 1 KATHERINE S. VIRGO, 1 JOSEPH T. D IPIRO,2 AVERY B. NATHENS, 3 ROBERT G. SAWYER, 4 and JOHN E.