Rangap1 Is a Continuous Marker of the Arabidopsis Cell Division Plane
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Investigating the Role of Cdk11in Animal Cytokinesis
Investigating the Role of CDK11 in Animal Cytokinesis by Thomas Clifford Panagiotou A thesis submitted in conformity with the requirements for the degree of Master of Science Department of Molecular Genetics University of Toronto © Copyright by Thomas Clifford Panagiotou (2020) Investigating the Role of CDK11 in Animal Cytokinesis Thomas Clifford Panagiotou Master of Science Department of Molecular Genetics University of Toronto 2020 Abstract Finely tuned spatio-temporal regulation of cell division is required for genome stability. Cytokinesis constitutes the final stages of cell division, from chromosome segregation to the physical separation of cells, abscission. Abscission is tightly regulated to ensure it occurs after earlier cytokinetic events, like the maturation of the stem body, the regulatory platform for abscission. Active Aurora B kinase enforces the abscission checkpoint, which blocks abscission until chromosomes have been cleared from the cytokinetic machinery. Currently, it is unclear how this checkpoint is overcome. Here, I demonstrate that the cyclin-dependent kinase CDK11 is required for cytokinesis. Both inhibition and depletion of CDK11 block abscission. Furthermore, the mitosis-specific CDK11p58 kinase localizes to the stem body, where its kinase activity rescues the defects of CDK11 depletion and inhibition. These results suggest a model whereby CDK11p58 antagonizes Aurora B kinase to overcome the abscission checkpoint to allow for successful completion of cytokinesis. ii Acknowledgments I am very grateful for the support of my family and friends throughout my studies. I would also like to express my deep gratitude to Wilde Lab members, both past and present, for their advice and collaboration. In particular, I am very grateful to Matthew Renshaw, whose work comprises part of this thesis. -

Phragmoplast Microtubule Dynamics – a Game of Zones Andrei Smertenko1,2,‡, Seanna L
© 2018. Published by The Company of Biologists Ltd | Journal of Cell Science (2018) 131, jcs203331. doi:10.1242/jcs.203331 REVIEW SPECIAL ISSUE: PLANT CELL BIOLOGY Phragmoplast microtubule dynamics – a game of zones Andrei Smertenko1,2,‡, Seanna L. Hewitt2,3, Caitlin N. Jacques2,4, Rafal Kacprzyk1, Yan Liu2,5, Matthew J. Marcec2,6, Lindani Moyo2,6, Aaron Ogden1,2, Hui Min Oung1,2, Sharol Schmidt1,2 and Erika A. Serrano-Romero2,5 ABSTRACT during anaphase from the remnants of the central spindle (Segui- Plant morphogenesis relies on the accurate positioning of the partition Simarro et al., 2004). It consists of microtubules, actin, membrane (cell plate) between dividing cells during cytokinesis. The cell plate is compartments and proteins that associate with or regulate the above synthetized by a specialized structure called the phragmoplast, which (Boruc and Van Damme, 2015; Lipka et al., 2015). The microtubule consists of microtubules, actin filaments, membrane compartments component of the phragmoplast consists of two aligned arrays that and associated proteins. The phragmoplast forms between daughter flank the so-called phragmoplast midzone, where cell plate assembly nuclei during the transition from anaphase to telophase. As cells are takes place (Fig. 1). The initial phragmoplast has a disk shape with a commonly larger than the originally formed phragmoplast, the diameter that approximately equals that of the daughter nuclei construction of the cell plate requires phragmoplast expansion. (Fig. 1); however, the parental cell is generally wider. For example, This expansion depends on microtubule polymerization at the the length of a cambium cell exceeds the diameter of the disk-shaped phragmoplast forefront (leading zone) and loss at the back (lagging phragmoplast during late anaphase by up to 100 fold (Larson, 1994). -

Mitosis Vs. Meiosis
Mitosis vs. Meiosis In order for organisms to continue growing and/or replace cells that are dead or beyond repair, cells must replicate, or make identical copies of themselves. In order to do this and maintain the proper number of chromosomes, the cells of eukaryotes must undergo mitosis to divide up their DNA. The dividing of the DNA ensures that both the “old” cell (parent cell) and the “new” cells (daughter cells) have the same genetic makeup and both will be diploid, or containing the same number of chromosomes as the parent cell. For reproduction of an organism to occur, the original parent cell will undergo Meiosis to create 4 new daughter cells with a slightly different genetic makeup in order to ensure genetic diversity when fertilization occurs. The four daughter cells will be haploid, or containing half the number of chromosomes as the parent cell. The difference between the two processes is that mitosis occurs in non-reproductive cells, or somatic cells, and meiosis occurs in the cells that participate in sexual reproduction, or germ cells. The Somatic Cell Cycle (Mitosis) The somatic cell cycle consists of 3 phases: interphase, m phase, and cytokinesis. 1. Interphase: Interphase is considered the non-dividing phase of the cell cycle. It is not a part of the actual process of mitosis, but it readies the cell for mitosis. It is made up of 3 sub-phases: • G1 Phase: In G1, the cell is growing. In most organisms, the majority of the cell’s life span is spent in G1. • S Phase: In each human somatic cell, there are 23 pairs of chromosomes; one chromosome comes from the mother and one comes from the father. -

The Revised Classification of Eukaryotes
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/231610049 The Revised Classification of Eukaryotes Article in Journal of Eukaryotic Microbiology · September 2012 DOI: 10.1111/j.1550-7408.2012.00644.x · Source: PubMed CITATIONS READS 961 2,825 25 authors, including: Sina M Adl Alastair Simpson University of Saskatchewan Dalhousie University 118 PUBLICATIONS 8,522 CITATIONS 264 PUBLICATIONS 10,739 CITATIONS SEE PROFILE SEE PROFILE Christopher E Lane David Bass University of Rhode Island Natural History Museum, London 82 PUBLICATIONS 6,233 CITATIONS 464 PUBLICATIONS 7,765 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Biodiversity and ecology of soil taste amoeba View project Predator control of diversity View project All content following this page was uploaded by Smirnov Alexey on 25 October 2017. The user has requested enhancement of the downloaded file. The Journal of Published by the International Society of Eukaryotic Microbiology Protistologists J. Eukaryot. Microbiol., 59(5), 2012 pp. 429–493 © 2012 The Author(s) Journal of Eukaryotic Microbiology © 2012 International Society of Protistologists DOI: 10.1111/j.1550-7408.2012.00644.x The Revised Classification of Eukaryotes SINA M. ADL,a,b ALASTAIR G. B. SIMPSON,b CHRISTOPHER E. LANE,c JULIUS LUKESˇ,d DAVID BASS,e SAMUEL S. BOWSER,f MATTHEW W. BROWN,g FABIEN BURKI,h MICAH DUNTHORN,i VLADIMIR HAMPL,j AARON HEISS,b MONA HOPPENRATH,k ENRIQUE LARA,l LINE LE GALL,m DENIS H. LYNN,n,1 HILARY MCMANUS,o EDWARD A. D. -

Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase
cells Review Role of Cyclin-Dependent Kinase 1 in Translational Regulation in the M-Phase Jaroslav Kalous *, Denisa Jansová and Andrej Šušor Institute of Animal Physiology and Genetics, Academy of Sciences of the Czech Republic, Rumburska 89, 27721 Libechov, Czech Republic; [email protected] (D.J.); [email protected] (A.Š.) * Correspondence: [email protected] Received: 28 April 2020; Accepted: 24 June 2020; Published: 27 June 2020 Abstract: Cyclin dependent kinase 1 (CDK1) has been primarily identified as a key cell cycle regulator in both mitosis and meiosis. Recently, an extramitotic function of CDK1 emerged when evidence was found that CDK1 is involved in many cellular events that are essential for cell proliferation and survival. In this review we summarize the involvement of CDK1 in the initiation and elongation steps of protein synthesis in the cell. During its activation, CDK1 influences the initiation of protein synthesis, promotes the activity of specific translational initiation factors and affects the functioning of a subset of elongation factors. Our review provides insights into gene expression regulation during the transcriptionally silent M-phase and describes quantitative and qualitative translational changes based on the extramitotic role of the cell cycle master regulator CDK1 to optimize temporal synthesis of proteins to sustain the division-related processes: mitosis and cytokinesis. Keywords: CDK1; 4E-BP1; mTOR; mRNA; translation; M-phase 1. Introduction 1.1. Cyclin Dependent Kinase 1 (CDK1) Is a Subunit of the M Phase-Promoting Factor (MPF) CDK1, a serine/threonine kinase, is a catalytic subunit of the M phase-promoting factor (MPF) complex which is essential for cell cycle control during the G1-S and G2-M phase transitions of eukaryotic cells. -

Molecular Biology and Applied Genetics
MOLECULAR BIOLOGY AND APPLIED GENETICS FOR Medical Laboratory Technology Students Upgraded Lecture Note Series Mohammed Awole Adem Jimma University MOLECULAR BIOLOGY AND APPLIED GENETICS For Medical Laboratory Technician Students Lecture Note Series Mohammed Awole Adem Upgraded - 2006 In collaboration with The Carter Center (EPHTI) and The Federal Democratic Republic of Ethiopia Ministry of Education and Ministry of Health Jimma University PREFACE The problem faced today in the learning and teaching of Applied Genetics and Molecular Biology for laboratory technologists in universities, colleges andhealth institutions primarily from the unavailability of textbooks that focus on the needs of Ethiopian students. This lecture note has been prepared with the primary aim of alleviating the problems encountered in the teaching of Medical Applied Genetics and Molecular Biology course and in minimizing discrepancies prevailing among the different teaching and training health institutions. It can also be used in teaching any introductory course on medical Applied Genetics and Molecular Biology and as a reference material. This lecture note is specifically designed for medical laboratory technologists, and includes only those areas of molecular cell biology and Applied Genetics relevant to degree-level understanding of modern laboratory technology. Since genetics is prerequisite course to molecular biology, the lecture note starts with Genetics i followed by Molecular Biology. It provides students with molecular background to enable them to understand and critically analyze recent advances in laboratory sciences. Finally, it contains a glossary, which summarizes important terminologies used in the text. Each chapter begins by specific learning objectives and at the end of each chapter review questions are also included. -

Functions of the Arabidopsis Kinesin Superfamily of Microtubule-Based Motor Proteins Chuanmei Zhu
Washington University in St. Louis Washington University Open Scholarship Biology Faculty Publications & Presentations Biology 10-2012 Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins Chuanmei Zhu Ram Dixit Washington University in St Louis, [email protected] Follow this and additional works at: https://openscholarship.wustl.edu/bio_facpubs Part of the Biochemistry Commons, Biology Commons, and the Plant Biology Commons Recommended Citation Zhu, Chuanmei and Dixit, Ram, "Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins" (2012). Biology Faculty Publications & Presentations. 79. https://openscholarship.wustl.edu/bio_facpubs/79 This Article is brought to you for free and open access by the Biology at Washington University Open Scholarship. It has been accepted for inclusion in Biology Faculty Publications & Presentations by an authorized administrator of Washington University Open Scholarship. For more information, please contact [email protected]. Functions of the Arabidopsis kinesin superfamily of microtubule-based motor proteins Chuanmei Zhu and Ram Dixit Biology Department, Washington University, St. Louis, MO 63130 Corresponding author Ram Dixit 1 Brookings Drive, CB 1137 St. Louis, MO 63130. Phone: (314) 935-8823 Fax: (314) 935-4432 Email: [email protected] Keywords Plant, cortical microtubule, preprophase band, spindle, phragmoplast ABSTRACT Plants possess a large number of microtubule-based kinesin motor proteins. While the Kinesin-2, 3, 9 and 11 families are absent from land plants, the Kinesin-7 and 14 families are greatly expanded. In addition, some kinesins are specifically present only in land plants. The distinctive inventory of plant kinesins suggests that kinesins have evolved to perform specialized functions in plants. -

List, Describe, Diagram, and Identify the Stages of Meiosis
Meiosis and Sexual Life Cycles Objective # 1 In this topic we will examine a second type of cell division used by eukaryotic List, describe, diagram, and cells: meiosis. identify the stages of meiosis. In addition, we will see how the 2 types of eukaryotic cell division, mitosis and meiosis, are involved in transmitting genetic information from one generation to the next during eukaryotic life cycles. 1 2 Objective 1 Objective 1 Overview of meiosis in a cell where 2N = 6 Only diploid cells can divide by meiosis. We will examine the stages of meiosis in DNA duplication a diploid cell where 2N = 6 during interphase Meiosis involves 2 consecutive cell divisions. Since the DNA is duplicated Meiosis II only prior to the first division, the final result is 4 haploid cells: Meiosis I 3 After meiosis I the cells are haploid. 4 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis Prophase I: ¾ Chromosomes condense. Because of replication during interphase, each chromosome consists of 2 sister chromatids joined by a centromere. ¾ Synapsis – the 2 members of each homologous pair of chromosomes line up side-by-side to form a tetrad consisting of 4 chromatids: 5 6 1 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis Prophase I: ¾ During synapsis, sometimes there is an exchange of homologous parts between non-sister chromatids. This exchange is called crossing over. 7 8 Objective 1, Stages of Meiosis Objective 1, Stages of Meiosis (2N=6) Prophase I: ¾ the spindle apparatus begins to form. ¾ the nuclear membrane breaks down: Prophase I 9 10 Objective 1, Stages of Meiosis Objective 1, 4 Possible Metaphase I Arrangements: Metaphase I: ¾ chromosomes line up along the equatorial plate in pairs, i.e. -

Cell Life Cycle and Reproduction the Cell Cycle (Cell-Division Cycle), Is a Series of Events That Take Place in a Cell Leading to Its Division and Duplication
Cell Life Cycle and Reproduction The cell cycle (cell-division cycle), is a series of events that take place in a cell leading to its division and duplication. The main phases of the cell cycle are interphase, nuclear division, and cytokinesis. Cell division produces two daughter cells. In cells without a nucleus (prokaryotic), the cell cycle occurs via binary fission. Interphase Gap1(G1)- Cells increase in size. The G1checkpointcontrol mechanism ensures that everything is ready for DNA synthesis. Synthesis(S)- DNA replication occurs during this phase. DNA Replication The process in which DNA makes a duplicate copy of itself. Semiconservative Replication The process in which the DNA molecule uncoils and separates into two strands. Each original strand becomes a template on which a new strand is constructed, resulting in two DNA molecules identical to the original DNA molecule. Gap 2(G2)- The cell continues to grow. The G2checkpointcontrol mechanism ensures that everything is ready to enter the M (mitosis) phase and divide. Mitotic(M) refers to the division of the nucleus. Cell growth stops at this stage and cellular energy is focused on the orderly division into daughter cells. A checkpoint in the middle of mitosis (Metaphase Checkpoint) ensures that the cell is ready to complete cell division. The final event is cytokinesis, in which the cytoplasm divides and the single parent cell splits into two daughter cells. Reproduction Cellular reproduction is a process by which cells duplicate their contents and then divide to yield multiple cells with similar, if not duplicate, contents. Mitosis Mitosis- nuclear division resulting in the production of two somatic cells having the same genetic complement (genetically identical) as the original cell. -

Growth Inhibition, Cytokinesis Failure and Apoptosis of Multidrug-Resistant
Leukemia (1999) 13, 768–778 1999 Stockton Press All rights reserved 0887-6924/99 $12.00 http://www.stockton-press.co.uk/leu Growth inhibition, cytokinesis failure and apoptosis of multidrug-resistant leukemia cells after treatment with P-glycoprotein inhibitory agents G Lehne1,2, P De Angelis3, M den Boer4 and HE Rugstad1 1Department of Clinical Pharmacology, 2Institute for Surgical Research, and 3Institute for Pathology, The National Hospital, Rikshospitalet, University of Oslo, Norway; and 4Department of Pediatrics, Free University Hospital, Amsterdam, The Netherlands The multidrug transporter P-glycoprotein (Pgp), which is fre- inhibitors with less toxic potential have been developed quently overexpressed in multidrug resistant leukemia, has including the non-immunosuppressive cyclosporin SDZ PSC many proposed physiological functions including involvement 833,12 the cyclopeptolide SDZ 280–446,13 and the cycloprop- in transmembraneous transport of certain growth-regulating 14 cytokines. Therefore, we studied cell growth of three pairs of yldibenzosuberane LY 335979. These agents exercise high drug resistant and sensitive leukemia cell lines (KG1a, K562 affinity for Pgp and interfere with binding of known substrates and HL60) exposed to three different inhibitors of Pgp. The such as azidopine and calcein.15–17 SDZ PSC 833 which is resistant KG1a and K562 sublines, which expressed high levels the most extensively studied of these, has proven to be well of Pgp, responded to low doses of the cyclosporin SDZ PSC tolerated in phase I trials,18–20 and is currently being investi- 833, the cyclopeptolide SDZ 280–446, and the cyclopropyldib- enzosuberane LY335979 with a dose-dependent growth inhi- gated as an adjunct to cancer chemotherapy in phase II and bition. -
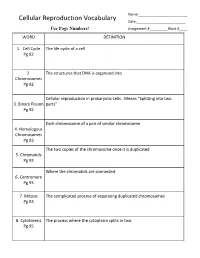
Cellular Reproduction Vocabulary Date:______Use Page Numbers! Assignment #______Block #____ WORD DEFINITION
Name:_________________________ Cellular Reproduction Vocabulary Date:_________________________ Use Page Numbers! Assignment #_________ Block #____ WORD DEFINITION 1. Cell Cycle The life cycle of a cell Pg 92 2. The structures that DNA is organized into Chromosomes Pg 92 Cellular reproduction in prokaryotic cells. Means “Splitting into two 3. Binary Fission parts” Pg 92 Each chromosome of a pair of similar chromosome 4. Homologous Chromosomes Pg 93 The two copies of the chromosome once it is duplicated 5. Chromatids Pg 93 Where the chromatids are connected 6. Centromere Pg 93 7. Mitosis The complicated process of separating duplicated chromosomes Pg 93 8. Cytokinesis The process where the cytoplasm splits in two Pg 95 WORD DEFINITION 9. Budding A type of asexual reproduction, where a piece of the parents body Pg 612 develops into an independent organism 10. A type of asexual reproduction, where the organism breaks into two or Regeneration more parts, each growing into a new organism that is genetically identical (fragmentation) to the parent Pg 612 The production of offspring by combining the genetic material of more 11. Sexual than one parent reproduction Pg 613 A single parent has offspring that are identical to itself 12. Asexual reproduction Pg 613 The male sex cell 13. Sperm Pg 613 The female sex cell 14. Egg Pg 613 The new type of cell that is made when an egg’s nucleus fuses with a 15. Zygote sperm’s nucleus Pg 613 16. Spores Small reproductive cells that are protected by a thick cell wall Pg 256 WORD DEFINITION 17. Sex Cells Specialized cells that combine to form a zygote, they have half the normal Pg 114 number of chromosomes, one of each pair. -

Meiosis I and Meiosis II; Life Cycles
Meiosis I and Meiosis II; Life Cycles Meiosis functions to reduce the number of chromosomes to one half. Each daughter cell that is produced will have one half as many chromosomes as the parent cell. Meiosis is part of the sexual process because gametes (sperm, eggs) have one half the chromosomes as diploid (2N) individuals. Phases of Meiosis There are two divisions in meiosis; the first division is meiosis I: the number of cells is doubled but the number of chromosomes is not. This results in 1/2 as many chromosomes per cell. The second division is meiosis II: this division is like mitosis; the number of chromosomes does not get reduced. The phases have the same names as those of mitosis. Meiosis I: prophase I (2N), metaphase I (2N), anaphase I (N+N), and telophase I (N+N) Meiosis II: prophase II (N+N), metaphase II (N+N), anaphase II (N+N+N+N), and telophase II (N+N+N+N) (Works Cited See) *3 Meiosis I (Works Cited See) *1 1. Prophase I Events that occur during prophase of mitosis also occur during prophase I of meiosis. The chromosomes coil up, the nuclear membrane begins to disintegrate, and the centrosomes begin moving apart. The two chromosomes may exchange fragments by a process called crossing over. When the chromosomes partially separate in late prophase, until they separate during anaphase resulting in chromosomes that are mixtures of the original two chromosomes. 2. Metaphase I Bivalents (tetrads) become aligned in the center of the cell and are attached to spindle fibers.