Atlantic-Mediterranean and Within- Mediterranean Molecular Variation in Coris Julis (L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Marine Biodiversity and Fisheries Catches of the Pitcairn Island Group
The Marine Biodiversity and Fisheries Catches of the Pitcairn Island Group THE MARINE BIODIVERSITY AND FISHERIES CATCHES OF THE PITCAIRN ISLAND GROUP M.L.D. Palomares, D. Chaitanya, S. Harper, D. Zeller and D. Pauly A report prepared for the Global Ocean Legacy project of the Pew Environment Group by the Sea Around Us Project Fisheries Centre The University of British Columbia 2202 Main Mall Vancouver, BC, Canada, V6T 1Z4 TABLE OF CONTENTS FOREWORD ................................................................................................................................................. 2 Daniel Pauly RECONSTRUCTION OF TOTAL MARINE FISHERIES CATCHES FOR THE PITCAIRN ISLANDS (1950-2009) ...................................................................................... 3 Devraj Chaitanya, Sarah Harper and Dirk Zeller DOCUMENTING THE MARINE BIODIVERSITY OF THE PITCAIRN ISLANDS THROUGH FISHBASE AND SEALIFEBASE ..................................................................................... 10 Maria Lourdes D. Palomares, Patricia M. Sorongon, Marianne Pan, Jennifer C. Espedido, Lealde U. Pacres, Arlene Chon and Ace Amarga APPENDICES ............................................................................................................................................... 23 APPENDIX 1: FAO AND RECONSTRUCTED CATCH DATA ......................................................................................... 23 APPENDIX 2: TOTAL RECONSTRUCTED CATCH BY MAJOR TAXA ............................................................................ -

Phylogeography of Indo‐Pacific Reef Fishes: Sister Wrasses Coris
Phylogeography of Indo-Pacific reef fishes: sister wrasses Coris gaimard and C. cuvieri in the Red Sea, Indian Ocean and Pacific Ocean Item Type Article Authors Ahti, Pauliina A.; Coleman, Richard R.; DiBattista, Joseph D.; Berumen, Michael L.; Rocha, Luiz A.; Bowen, Brian W. Citation Ahti, P. A., Coleman, R. R., DiBattista, J. D., Berumen, M. L., Rocha, L. A., & Bowen, B. W. (2016). Phylogeography of Indo-Pacific reef fishes: sister wrassesCoris gaimardandC. cuvieriin the Red Sea, Indian Ocean and Pacific Ocean. Journal of Biogeography, 43(6), 1103–1115. doi:10.1111/jbi.12712 Eprint version Post-print DOI 10.1111/jbi.12712 Publisher Wiley Journal Journal of Biogeography Rights Archived with thanks to Wiley; This file is an open access version redistributed from: https://rss.onlinelibrary.wiley.com/doi/am- pdf/10.1111/jbi.12712 Download date 23/09/2021 11:07:08 Link to Item http://hdl.handle.net/10754/621458 1 Received Date : 13-Dec-2014 2 Revised Date : 14-Nov-2015 3 Accepted Date : 09-Dec-2015 4 Article type : Special Issue 5 Original Article 6 LRH: P. A. Ahti et al. 7 RRH: Phylogeography of Indo-Pacific coral reef fishes 8 Phylogeography of Indo-Pacific reef fishes: sister wrasses Coris gaimard and 9 C. cuvieri in the Red Sea, Indian Ocean and Pacific Ocean 10 Pauliina A. Ahti1,2*, Richard R. Coleman1, Joseph D. DiBattista3,4, Michael L. Berumen3, Luiz A. 11 Rocha5 and Brian W. Bowen1 12 1Hawai‘i Institute of Marine Biology, University of Hawai‘i, Kāne‘ohe, HI 96744, USA, 2School 13 of Life Sciences, College of Medical, Veterinary and Life Sciences, University of Glasgow, 14 Glasgow, G12 8QQ, Scotland, 3Red Sea Research Center, Division of Biological and 15 Environmental Science and Engineering, King Abdullah University of Science and Technology 16 (KAUST), Thuwal, 23955-6900, Saudi Arabia, 4Department of Environment and Agriculture, 17 Curtin University, PO Box U1987, Perth, WA 6845, Australia , 5Section of Ichthyology, 18 California Academy of Sciences, San Francisco, CA 94118, USA 19 *Correspondence: Pauliina A. -

What Explains the Diversity of Sexually Selected Traits?
Biol. Rev. (2020), 95, pp. 847–864. 847 doi: 10.1111/brv.12593 Songs versus colours versus horns: what explains the diversity of sexually selected traits? John J. Wiens* and E. Tuschhoff Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, AZ, 85721-0088, U.S.A. ABSTRACT Papers on sexual selection often highlight the incredible diversity of sexually selected traits across animals. Yet, few studies have tried to explain why this diversity evolved. Animals use many different types of traits to attract mates and outcom- pete rivals, including colours, songs, and horns, but it remains unclear why, for example, some taxa have songs, others have colours, and others horns. Here, we first conduct a systematic survey of the basic diversity and distribution of dif- ferent types of sexually selected signals and weapons across the animal Tree of Life. Based on this survey, we describe seven major patterns in trait diversity and distributions. We then discuss 10 unanswered questions raised by these pat- terns, and how they might be addressed. One major pattern is that most types of sexually selected signals and weapons are apparently absent from most animal phyla (88%), in contrast to the conventional wisdom that a diversity of sexually selected traits is present across animals. Furthermore, most trait diversity is clustered in Arthropoda and Chordata, but only within certain clades. Within these clades, many different types of traits have evolved, and many types appear to have evolved repeatedly. By contrast, other major arthropod and chordate clades appear to lack all or most trait types, and similar patterns are repeated at smaller phylogenetic scales (e.g. -

Shallow Water Records of Fish
Shallow water records of fish Collected from or observed at Howland Island from 1927-2002. Collected or compiled by Mundy et al. (2002). Scientific Name Common Name GINGLYMOSTOMATIDAE Nurse Sharks Nebrius ferrugineus nurse shark CARCHARHINIDAE Requiem Sharks Carcharhinus amblyrhynchos grey reef shark Carcharhinus melanopterus reef blacktip shark Galeocerdo cuvieri tiger shark HEMIGALEIDAE Weasel Sharks Triaenodon obesus reef whitetip shark SPHYRNIDAE Hammerhead Sharks Sphyrna lewini scalloped hammerhead shark DASYATIDAE Sand Rays Taeniura meyeni MYLIOBATIDIDAE Eagle Rays Aetobatus narinari spotted eagle ray MOBULIDAE Manta Rays Manta sp. manta MURAENIDAE Moray Eels Anarchias allardicei Allardice’s moray Anarchias cantonenesis Canton Island moray Echidna nebulosa snowflake moray Echidna polyzona barred moray Enchelycore pardalis Gymnomuraena zebra zebra moray Gymnothorax breedini Gymnothorax chilospilus Gymnothorax javanicus giant moray Gymnothorax flavimarginatus yellow-margined moray Gymnothorax marshallensis Marshall Island moray Gymnothorax meleagris white-mouth moray Gymnothorax picta peppered moray Gymnothorax rueppelliae yellow-headed moray Gymnothorax sp. Gymnothorax thyrsoideus Gymnothorax undulatus undulated moray Uropterygius sp. Uropterygius marmoratus marbled snake moray OPHICHTHIDAE Myrichthys maculosus spotted snake eel CONGRIDAE Conger Eel Conger sp. CHANIDAE Milkfish Chanos chanos milkfish SYNODONTIDAE Lizardfishes Synodus sp. HOLOCENTRIDAE Squirrelfishes Myripristis berndti bigscale soldierfish Sargocentron caudimaculatum -

PDF Download
A checklist of wrasses (Labridae) and parrotfishes (Scaridae) of the world: 2017 update PAOLO PARENTI Department of Earth and Environmental Sciences, University of Milano-Bicocca, Piazza della Scienza 1, 20126 Milano, Italy E-mail: [email protected] JOHN E. RANDALL Bishop Museum, 1525 Bernice St., Honolulu, HI 96817-2704, USA E-mail: [email protected] Abstract The wrasse family Labridae sensu stricto contains 548 species at latest count at the end of 2017. The previous checklist by Parenti & Randall (2011) included 504 species: the additional species represent new discoveries (41 new species) plus resurrections from synonymy (6) minus species names synonymized (3). The total number of genera is maintained at 70 as two new genera have been erected (Sagittalarva Victor, Alfaro & Sorenson, 2013 for Pseudojuloides inornatus (Gilbert, 1890) and Novaculops Schultz, 1960, removed from synonymy with Xyrichtys Cuvier, 1814), and two genera have been synonymized: Nelabrichthys Russell, 1983 is a junior synonym of Suezichthys Smith, 1958 (Russell & Westneat 2013) and Xiphocheilus Bleeker, 1856 is now regarded as a subgenus of Choerodon Bleeker, 1847 (Gomon 2017). There are some discrepancies with the total number of labrid species (558) reported by the Catalog of Fishes by Eschmeyer et al. (2018) and they are discussed herein. Only one species of parrotfish (Scaridae) has been described as new since the last checklist in 2011. Key words: ichthyology, taxonomy, systematics, coral reef fishes, species list, catalog, Indo-Pacific Ocean. Citation: Parenti, P. & Randall, J.E. (2018) A checklist of wrasses (Labridae) and parrotfishes (Scaridae) of the world: 2017 update. Journal of the Ocean Science Foundation, 30, 11–27. -

Phylogeography of Indo-Pacific Reef Fishes: Sister Wrasses Coris Gaimard And
This is the peer reviewed version of the article which has been published in final form at http://doi.org/10.1111/jbi.12712. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Self-Archiving at http://olabout.wiley.com/WileyCDA/Section/id-828039.html#terms 1 1 For the virtual issue, "Red Sea and Western Indian Ocean Biogeography" 2 Original Article 3 LRH: P. A. Ahti et al. 4 RRH: Phylogeography of Indo-Pacific coral reef fishes 5 Phylogeography of Indo-Pacific reef fishes: Sister wrasses Coris gaimard and 6 C. cuvieri in the Red Sea, Indian Ocean, and Pacific Ocean 7 Pauliina A. Ahti1,2*, Richard R. Coleman1, Joseph D. DiBattista3,4, Michael L. Berumen3, Luiz A. 8 Rocha5 and Brian W. Bowen1 9 1Hawai‘i Institute of Marine Biology, University of Hawai‘i, Kāne‘ohe, HI 96744, USA, 2School 10 of Life Sciences, College of Medical, Veterinary and Life Sciences, University of Glasgow, 11 Glasgow, G12 8QQ, Scotland, 3Red Sea Research Center, Division of Biological and 12 Environmental Science and Engineering, King Abdullah University of Science and Technology 13 (KAUST), Thuwal, 23955-6900, Saudi Arabia, 4Department of Environment and Agriculture, 14 Curtin University, PO Box U1987, Perth, WA 6845, Australia , 5Section of Ichthyology, 15 California Academy of Sciences, San Francisco, CA 94118, USA 16 *Correspondence: Pauliina A. Ahti, School of Life Sciences, College of Medical, Veterinary 17 and Life Sciences, University of Glasgow, Glasgow, G12 8QQ, Scotland, E-mail: 18 [email protected] 19 20 Word count: 7454 2 21 ABSTRACT 22 Aim We aim to resolve evolutionary history, biogeographic barriers and population histories 23 for sister species of wrasses, the African Coris (Coris cuvieri) in the Indian Ocean and Red 24 Sea, and the Yellowtail Coris (Coris gaimard) in the Pacific Ocean. -

Vertebrates - Advanced
Vertebrates - Advanced Douglas Wilkin, Ph.D. Jennifer Blanchette Niamh Gray-Wilson Jean Brainard, Ph.D. Say Thanks to the Authors Click http://www.ck12.org/saythanks (No sign in required) AUTHORS Douglas Wilkin, Ph.D. To access a customizable version of this book, as well as other Jennifer Blanchette interactive content, visit www.ck12.org Niamh Gray-Wilson Jean Brainard, Ph.D. EDITOR Douglas Wilkin, Ph.D. CK-12 Foundation is a non-profit organization with a mission to reduce the cost of textbook materials for the K-12 market both in the U.S. and worldwide. Using an open-source, collaborative, and web-based compilation model, CK-12 pioneers and promotes the creation and distribution of high-quality, adaptive online textbooks that can be mixed, modified and printed (i.e., the FlexBook® textbooks). Copyright © 2016 CK-12 Foundation, www.ck12.org The names “CK-12” and “CK12” and associated logos and the terms “FlexBook®” and “FlexBook Platform®” (collectively “CK-12 Marks”) are trademarks and service marks of CK-12 Foundation and are protected by federal, state, and international laws. Any form of reproduction of this book in any format or medium, in whole or in sections must include the referral attribution link http://www.ck12.org/saythanks (placed in a visible location) in addition to the following terms. Except as otherwise noted, all CK-12 Content (including CK-12 Curriculum Material) is made available to Users in accordance with the Creative Commons Attribution-Non-Commercial 3.0 Unported (CC BY-NC 3.0) License (http://creativecommons.org/ licenses/by-nc/3.0/), as amended and updated by Creative Com- mons from time to time (the “CC License”), which is incorporated herein by this reference. -

Fanny WITKOWSKI*, Aurélie VION, and Marc BOUCHOUCHA
ACTA ICHTHYOLOGICA ET PISCATORIA (2016) 46 (3): 171–183 DOI: 10.3750/AIP2016.46.3.02 TEMPORAL PARTITIONING OF DIURNAL BEHAVIOURAL PATTERNS OF CORIS JULIS AND DIPLODUS VULGARIS (ACTINOPTERYGII: PERCIFORMES) IN MEDITERRANEAN CORALLIGENOUS HABITATS Fanny WITKOWSKI*, Aurélie VION, and Marc BOUCHOUCHA Ifremer, Laboratoire Environnement Ressources Provence Azur Corse, Zone portuaire de Brégaillon, 83507 La Seyne-sur-Mer, France Witkowski F., Vion A., Bouchoucha M. 2016. Temporal partitioning of diurnal behavioural patterns of Coris julis and Diplodus vulgaris (Actinopterygii: Perciformes) in Mediterranean coralligenous habitats. Acta Ichthyol. Piscat. 46 (3): 171–183. Background. There is need for more information on behavioural rhythms of fi shes in relation to their habitat use in few or not previously monitored areas, such as coralligenous habitats. Some studies have compiled inventories of observable fi sh species in this habitat, but no studies have been performed on the temporal patterning of fi sh behaviour, especially at the scale of the day. The presently reported study aims at providing knowledge on fi ne- scale fl uctuations of abundances and behaviour of two fi sh species in coralligenous habitats throughout the diurnal period. Materials and methods. Fish fauna was fi lmed hourly, for several days, from dusk to dawn, with autonomous, programmable, and rotating video systems. Diurnal variations in abundances per count, considering size classes and behaviour, were studied for two diurnal fi sh species— the Mediterranean rainbow wrasse, Coris julis (Linnaeus, 1758) (Labridae), and the common two-banded seabream, Diplodus vulgaris (Geoffroy Saint-Hilaire, 1817) (Sparidae) in two study areas of coralligenous habitats on the French Riviera. Results. -
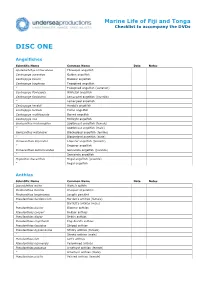
Fiji and Tonga Checklist to Accompany the Dvds
Marine Life of Fiji and Tonga Checklist to accompany the DVDs DISC ONE Angelfishes Scientific Name Common Name Date Notes Apolemichthys trimaculatus Threespot angelfish Centropyge aurantius Golden angelfish Centropyge bicolor Bicolour angelfish Centropyge bispinosa Twospined angelfish ” Twospined angelfish (variation) Centropyge flavicauda Whitetail angelfish Centropyge flavissima Lemonpeel angelfish (juvenile) ” Lemonpeel angelfish Centropyge heraldi Herald's angelfish Centropyge loricula Flame angelfish Centropyge multifasciata Barred angelfish Centropyge nox Midnight angelfish Genicanthus melanospilos Spotbreast angelfish (female) ” Spotbreast angelfish (male) Genicanthus watanabei Blackedged angelfish (female) ” Blackedged angelfish (male) Pomacanthus imperator Emperor angelfish (juvenile) ” Emperor angelfish Pomacanthus semicirculatus Semicircle angelfish (juvenile) ” Semicircle angelfish Pygoplites diacanthus Regal angelfish (juvenile) ” Regal angelfish Anthias Scientific Name Common Name Date Notes Luzonichthys waitei Waite's splitfin Plectranthias inermis Chequered perchlet Plectranthias longimanus Longfin perchlet Pseudanthias bartlettorum Bartlett's anthias (female) ” Bartlett's anthias (male) Pseudanthias bicolor Bicolour anthias Pseudanthias cooperi Redbar anthias Pseudanthias dispar Redfin anthias Pseudanthias engelhardi Engelhard's anthias Pseudanthias fasciatus Striped anthias Pseudanthias hypselosoma Stocky anthias (female) ” Stocky anthias (male) Pseudanthias lori Lori's anthias Pseudanthias luzonensis Yellowlined anthias -

The First Fish Field Guide Ever Published Dedicated to the Tubbataha Reefs
The first fish field guide ever published dedicated to the Tubbataha Reefs Compiled for the use of guests, visitors, and marine scientists With over 200 pictures of different fish species Documented >30% of the total resident population (633) TUBBATAHA REEFS FISH GUIDE TUBBATAHA REEFS FISH GUIDE Acknowledgements Author’s note Foreword Acanthuridae Surgeonfishes, Unicornfishes (p. 1) Aulostomidae Trumpetfishes (p. 5) Balistidae Triggerfishes (p. 6) Caesionidae Fusiliers (p. 8) Carangidae Jacks, Trevallies (p. 9) Chaetodontidae Butterflyfishes (p. 10) Cirrhitidae Hawkfishes (p. 14) Ephippidae Batfishes (p. 15) Fistularidae Cornetfishes (p. 16) Gobiidae Gobies (p. 17) Sweetlips (p. 18) Haemulidae Holocentridae Squirrelfishes (p. 19) Kyphosidae Sea chubs (p. 20) Labridae Wrasses (p. 21) Lethrinidae Emperors (p. 26) Lutjanidae Snappers (p. 29) Mullidae Goatfishes (p. 32) Muraenidae Moray eels (p. 33) Nemipteridae Whiptail breams (p. 34) Ostraciidae Boxfishes (p. 35) Pinguipedidae Sand perches (p. 36) Pomacanthidae Angelfishes (p. 37) Pomacentridae Damselfishes (p. 38) Priacanthidae Bigeyes (p. 44) Ptereleotridae Dartfishes (p. 45) Scaridae Parrotfishes (p. 46) Scorpaenidae Scorpionfishes, Firefishes (p. 49) Serranidae Groupers, Sea basses (p. 50) Siganidae Rabbitfishes (p. 54) Sphyraenidae Barracudas (p. 55) Synodontidae Lizardfishes (p. 56) Tetraodontidae Pufferfishes (p. 57) Zanclidae Moorish idol (p. 59) FAMILY ACANTHURIDAE Surgeonfishes, Tangs, Unicornfishes from Greek Akantha Class ACTINOPTERYGII (thorn) + Oura (tail) Subclass Order PERCIFORMES Suborder ACANTHUROIDEI REMARKS - All species marine, circumtropical distribution, 82 species recorded under 6 genera - Small and terminal mouth, ctenoid scales, single row close-set teeth, many are brightly colored - Feed on benthic algae, zooplankton, and detritus. Pelagic spawners © Toppx2 © Segundo Conales Jr. 2013 JAPANESE SURGEONFISH LINED SURGEONFISH Acanthurus japonicus Acanthurus lineatus Max. -

Family-Group Names of Recent Fishes
Zootaxa 3882 (2): 001–230 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Monograph ZOOTAXA Copyright © 2014 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3882.1.1 http://zoobank.org/urn:lsid:zoobank.org:pub:03E154FD-F167-4667-842B-5F515A58C8DE ZOOTAXA 3882 Family-group names of Recent fishes RICHARD VAN DER LAAN1,5, WILLIAM N. ESCHMEYER2 & RONALD FRICKE3,4 1Grasmeent 80, 1357JJ Almere, The Netherlands. E-mail: [email protected] 2Curator Emeritus, California Academy of Sciences, 55 Music Concourse Drive, San Francisco, CA 94118, USA. E-mail: [email protected] 3Im Ramstal 76, 97922 Lauda-Königshofen, Germany. E-mail: [email protected] 4Staatliches Museum für Naturkunde Stuttgart, Rosenstein 1, D-70191 Stuttgart, Germany [temporarily out of office] 5Corresponding author Magnolia Press Auckland, New Zealand Accepted by L. Page: 6 Sept. 2014; published: 11 Nov. 2014 Licensed under a Creative Commons Attribution License http://creativecommons.org/licenses/by/3.0 RICHARD VAN DER LAAN, WILLIAM N. ESCHMEYER & RONALD FRICKE Family-group names of Recent fishes (Zootaxa 3882) 230 pp.; 30 cm. 11 Nov. 2014 ISBN 978-1-77557-573-3 (paperback) ISBN 978-1-77557-574-0 (Online edition) FIRST PUBLISHED IN 2014 BY Magnolia Press P.O. Box 41-383 Auckland 1346 New Zealand e-mail: [email protected] http://www.mapress.com/zootaxa/ © 2014 Magnolia Press 2 · Zootaxa 3882 (1) © 2014 Magnolia Press VAN DER LAAN ET AL. Table of contents Abstract . .3 Introduction . .3 Methods . .5 Rules for the family-group names and how we dealt with them . .6 How to use the family-group names list . -
Glossary of Terminology
NOAA Coral Reef Information System - Glossary of Terminology Coral Reef Information System Home Data & Publications Regional Portals CRCP Activities Glossary HOME> GLOSSARY HOME> GLOSSARY OF TERMINOLOGY AND ACRONYMS/ABBREVIATIONS Glossary of Terminology A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | abalone a univalve mollusk (class Gastropoda) of the genus Haliotis. Abalones are harvested commercially for food consumption. The shell is lined with mother- of-pearl and used for commercial (ornamental) purposes Sea otters are in direct competition with humans for abalone. (Photo: Ron McPeak) abatement reducing the degree or intensity of, or eliminating abaxial away from, or distant from the axis abbreviate shortened abdomen in higher animals, the portion of the body that contains the intestines and other http://www.coris.noaa.gov/glossary/print-glossary.html[3/10/2016 11:56:15 AM] NOAA Coral Reef Information System - Glossary of Terminology viscera other than the lungs and heart; in arthropods, the rearmost segment of the body, which contains part of the digestive tract and all the reproductive organs The ventral surface of the abdomen of an American lobster. Prominent are the swimmerettes, uropods, and telson. abdominal fin a term used to describe the location of the pelvic (ventral) fins when they are inserted far behind pectorals. This is the more primitive condition. More recently evolved conditions have the pelvic fins in the thoracic or jugular positions. A salmon, for example, has its pelvic fins in the abdominal position. An angelfish has the pelvic fins in the thoracic position, and blennies have the pelvic fins in the jugular position, anterior to the pelvic girdle abductor a type of muscle whose function is to move an appendage or body part away from the body of an animal.