TRACE: Tennessee Research and Creative Exchange
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
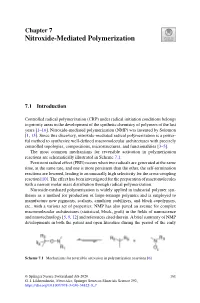
Nitroxide-Mediated Polymerization
Chapter 7 Nitroxide-Mediated Polymerization 7.1 Introduction Controlled radical polymerization (CRP) under radical initiation conditions belongs to priority areas in the development of the synthetic chemistry of polymers of the last years [1–16]. Nitroxide-mediated polymerization (NMP) was invented by Solomon [1, 13]. Since this discovery, nitroxide-mediated radical polymerization is a power- ful method to synthesize well-defined macromolecular architectures with precisely controlled topologies, compositions, microstructures, and functionalities [3–5]. The most common mechanisms for reversible activation in polymerization reactions are schematically illustrated in Scheme 7.1. Persistent radical effect (PRE) occurs when two radicals are generated at the same time, at the same rate, and one is more persistent than the other, the self-termination reactions are lowered, leading to an unusually high selectivity for the cross-coupling reaction [10]. The effect has been investigated for the preparation of macromolecules with a narrow molar mass distribution through radical polymerization. Nitroxide-mediated polymerization is widely applied in industrial polymer syn- theses as a method for production of large-tonnage polymers and is employed to manufacture new pigments, sealants, emulsion stabilizers, and block copolymers, etc., with a various set of properties. NMP has also paved an avenue for complex macromolecular architectures (statistical, block, graft) in the fields of nanoscience and nanotechnology [5, 9, 12] and references cited therein. A brief summary of NMP developments in both the patent and open literature during the period of the early Scheme 7.1 Mechanisms for reversible activation in polymerization reactions [6] © Springer Nature Switzerland AG 2020 161 G. I. Likhtenshtein, Nitroxides, Springer Series in Materials Science 292, https://doi.org/10.1007/978-3-030-34822-9_7 162 7 Nitroxide-Mediated Polymerization 1980–2000 was presented in [11]. -

Exosomes Confer Chemoresistance to Pancreatic Cancer Cells By
FULL PAPER British Journal of Cancer (2017) 116, 609–619 | doi: 10.1038/bjc.2017.18 Keywords: chemoresistance; exosomes; pancreatic cancer; ROS; microRNA Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK Girijesh Kumar Patel1, Mohammad Aslam Khan1, Arun Bhardwaj1, Sanjeev K Srivastava1, Haseeb Zubair1, Mary C Patton1, Seema Singh1,2, Moh’d Khushman3 and Ajay P Singh*,1,2 1Department of Oncologic Sciences, Mitchell Cancer Institute, University of South Alabama, Mobile, AL, USA; 2Department of Biochemistry and Molecular Biology, College of Medicine, University of South Alabama, Mobile, AL, USA and 3Department of Interdisciplinary Clinical Oncology, Mitchell Cancer Institute, University of South Alabama, Mobile, AL, USA Background: Chemoresistance is a significant clinical problem in pancreatic cancer (PC) and underlying molecular mechanisms still remain to be completely understood. Here we report a novel exosome-mediated mechanism of drug-induced acquired chemoresistance in PC cells. Methods: Differential ultracentrifugation was performed to isolate extracellular vesicles (EVs) based on their size from vehicle- or gemcitabine-treated PC cells. Extracellular vesicles size and subtypes were determined by dynamic light scattering and marker profiling, respectively. Gene expression was examined by qRT-PCR and/or immunoblot analyses, and direct targeting of DCK by miR-155 was confirmed by dual-luciferase 30-UTR reporter assay. Flow cytometry was performed to examine the apoptosis indices and reactive oxygen species (ROS) levels in PC cells using specific dyes. Cell viability was determined using the WST-1 assay. Results: Conditioned media (CM) from gemcitabine-treated PC cells (Gem-CM) provided significant chemoprotection to subsequent gemcitabine toxicity and most of the chemoresistance conferred by Gem-CM resulted from its EVs fraction. -

The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (Cad) by Phosphorylation and Protein-Protein Interactions
THE REGULATION OF CARBAMOYL PHOSPHATE SYNTHETASE-ASPARTATE TRANSCARBAMOYLASE-DIHYDROOROTASE (CAD) BY PHOSPHORYLATION AND PROTEIN-PROTEIN INTERACTIONS Eric M. Wauson A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology. Chapel Hill 2007 Approved by: Lee M. Graves, Ph.D. T. Kendall Harden, Ph.D. Gary L. Johnson, Ph.D. Aziz Sancar M.D., Ph.D. Beverly S. Mitchell, M.D. 2007 Eric M. Wauson ALL RIGHTS RESERVED ii ABSTRACT Eric M. Wauson: The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (CAD) by Phosphorylation and Protein-Protein Interactions (Under the direction of Lee M. Graves, Ph.D.) Pyrimidines have many important roles in cellular physiology, as they are used in the formation of DNA, RNA, phospholipids, and pyrimidine sugars. The first rate- limiting step in the de novo pyrimidine synthesis pathway is catalyzed by the carbamoyl phosphate synthetase II (CPSase II) part of the multienzymatic complex Carbamoyl phosphate synthetase, Aspartate transcarbamoylase, Dihydroorotase (CAD). CAD gene induction is highly correlated to cell proliferation. Additionally, CAD is allosterically inhibited or activated by uridine triphosphate (UTP) or phosphoribosyl pyrophosphate (PRPP), respectively. The phosphorylation of CAD by PKA and ERK has been reported to modulate the response of CAD to allosteric modulators. While there has been much speculation on the identity of CAD phosphorylation sites, no definitive identification of in vivo CAD phosphorylation sites has been performed. Therefore, we sought to determine the specific CAD residues phosphorylated by ERK and PKA in intact cells. -

Corynebacterium Glutamicum Mycoredoxin 3 Protects Against Multiple Oxidative Stresses
Advance Publication J. Gen. Appl. Microbiol. doi 10.2323/jgam.2019.10.003 ©2020 Applied Microbiology, Molecular and Cellular Biosciences Research Foundation Full Paper 1 Corynebacterium glutamicum Mycoredoxin 3 protects against multiple oxidative stresses 2 and displays thioredoxin-like activity 3 (Received September 24, 2019; Accepted October 28, 2019; J-STAGE Advance publication date: October 30, 2020) 4 Tao Su#, Chengchuan Che#, Ping Sun, Xiaona Li, Zhijin Gong, Jinfeng Liu, Ge Yang * 5 6 College of Life Sciences, Qufu Normal University, Qufu, Shandong 273165, China; 7 8 Running title: Feature of C. glutamicum mycoredoxin 3 9 10 11 # These authors contributed equally to this work. 12 13 * Corresponding authors: 14 Ge Yang 15 16 E-mail [email protected] 17 Tel: 86-13953760056 18 19 20 21 22 23 24 25 26 27 Abstract 28 Glutaredoxins (Grxs) and thioredoxins (Trxs) play a critical role in resistance to oxidative 29 conditions. However, physiological and biochemical roles of Mycoredoxin 3 (Mrx3) that shared a 30 high amino acid sequence similarity to Grxs remain unknown in Corynebacterium glutamicum. 31 Here we showed that mrx3 deletion strains of C. glutamicum was involved in the protection 32 against oxidative stress. Recombinant Mrx3 not only catalytically reduced the disulfide bonds in 33 ribonucleotide reductase (RNR), insulin and 5, 5’-dithiobis-(2-nitro-benzoicacid) (DTNB), but 34 also reduced the mixed disulphides between mycothiol (MSH) and substrate, which was 35 exclusively linked to the thioredoxin reductase (TrxR) electron transfer pathway by a dithiol 36 mechanism. Site-directed mutagenesis confirmed that the conserved Cys17 and Cys20 in Mrx3 37 were necessary to maintain its activity. -
Generate Metabolic Map Poster
Authors: Peter D. Karp Suzanne Paley Julio Collado-Vides John L Ingraham Ingrid Keseler Markus Krummenacker Cesar Bonavides-Martinez Robert Gunsalus An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Carol Fulcher Ian Paulsen Socorro Gama-Castro Robert LaRossa Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. EcoCyc: Escherichia coli K-12 substr. MG1655 Cellular Overview Connections between pathways are omitted for legibility. Anamika Kothari Amanda Mackie Alberto Santos-Zavaleta succinate phosphate succinate N-acetyl-DL-methionine + L-ornithine glutathione + L-methionine S-oxide D-fructofuranose γ Ag+ molybdate ferroheme b L,L-homocystine asp lys cys L-alanyl- -D- D-mannopyranose 6-phosphate 2+ 2+ H D-methionine 2-deoxy-D-glucose succinate formate formate succinate D-tartrate putrescine agmatine cadaverine L-tartrate D-fructofuranose 6-phosphate + nitrate nitrate Cu thiosulfate deoxycholate L,L-homocystine D-cystine D-cycloserine methyl β-D-glucoside putrescine asp spermidine (S)-2-hydroxybutanoate (S)-2-hydroxybutanoate arg L-homoserine lactone magnesium hydrogenphosphate magnesium hydrogenphosphate antimonous acid glutamyl-meso- Co2+ Cd2+ lactulose poly-β-1,6- met cob(I)inamide 2,3-dioxo- -

Ribonucleotide Reductase Subunit M2B Deficiency Leads To
Am J Transl Res 2018;10(11):3635-3649 www.ajtr.org /ISSN:1943-8141/AJTR0084684 Original Article Ribonucleotide reductase subunit M2B deficiency leads to mitochondrial permeability transition pore opening and is associated with aggressive clinicopathologic manifestations of breast cancer Lijun Xue1*, Xiyong Liu2,6*, Qinchuan Wang3,4, Charlie Q Liu3, Yunru Chen3, Wei Jia5, Ronhong Hsie6, Yifan Chen8, Frank Luh2,6, Shu Zheng7, Yun Yen2,6,8 1Department of Pathology, Loma Linda University Medical Center, Loma Linda, CA 92354, USA; 2Sino-American Cancer Foundation, California Cancer Institute, Temple, CA 91780, USA; 3Department of Molecular Pharmacology, Beckman Research Institute, City of Hope Comprehensive Cancer Center, Duarte, CA 91010, USA; 4Surgical Oncology, Sir Runrun Shaw Hospital, School of Medicine, Zhejiang University, Hangzhou, Zhejiang, China; 5Cancer Epidemiology Program, University of Hawaii Cancer Center, Honolulu, HI 96813, USA; 6TMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei, Taiwan, ROC; 7Cancer Institute, Zhejiang University, Hangzhou 310009, Zhejiang, China; 8PhD Program of Cancer Biology and Drug Discovery, Taipei Medical University, Taipei, Taiwan, ROC. *Co-first authors. Received August 27, 2018; Accepted October 19, 2018; Epub November 15, 2018; Published November 30, 2018 Abstract: Ribonucleotide reductase small subunit M2B (RRM2B) plays an essential role in maintaining mitochon- drial homeostasis. Mitochondrial permeability transition pore (MPTP) is a key regulator of mitochondrial homeo- stasis. MPTP contributes to cell death and is crucial in cancer progression. RRM2B’s relation to MPTP is not well known, and the role of RRM2B in cancer progression is controversial. Here, our aim was to study the role of RRM2B in regulating MPTP and the association between RRM2B and clinicopathological manifestations in breast cancer. -

The Role of Intermembrane Space Redox Factors in Glutathione Metabolism and Intracellular Redox Equilibrium Hatice Kubra Ozer University of South Carolina
University of South Carolina Scholar Commons Theses and Dissertations 2015 The Role of Intermembrane Space Redox Factors In Glutathione Metabolism And Intracellular Redox Equilibrium Hatice Kubra Ozer University of South carolina Follow this and additional works at: https://scholarcommons.sc.edu/etd Part of the Chemistry Commons Recommended Citation Ozer, H. K.(2015). The Role of Intermembrane Space Redox Factors In Glutathione Metabolism And Intracellular Redox Equilibrium. (Doctoral dissertation). Retrieved from https://scholarcommons.sc.edu/etd/3702 This Open Access Dissertation is brought to you by Scholar Commons. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. THE ROLE OF INTERMEMBRANE SPACE REDOX FACTORS IN GLUTATHIONE METABOLISM AND INTRACELLULAR REDOX EQUILIBRIUM by Hatice Kubra Ozer Bachelor of Science Uludag University, 2004 Master of Food Science and Nutrition Clemson University, 2010 Submitted in Partial Fulfillment of the Requirements For the Degree of Doctor of Philosophy in Chemistry College of Arts and Sciences University of South Carolina 2015 Accepted by: Caryn E. Outten, Major Professor F. Wayne Outten, Committee Chair Erin Connolly, Committee Member Andrew B. Greytak, Committee Member Lacy K. Ford, Senior Vice Provost and Dean of Graduate Studies © Copyright by Hatice Kubra Ozer, 2015 All Rights Reserved. ii ACKNOWLEDGEMENTS First, I would like to thank my advisor, Dr. Caryn E. Outten for her patience, guidance, expertise, and confidence in me to complete the work contained herein. She has been an excellent mentor during my graduate program. She is also the only person beside myself who is guaranteed to have read every word of this manuscript and to review every presentations in the past and her insight was irreplaceable. -

Role of GSH and Iron-Sulfur Glutaredoxins in Iron Metabolism—Review
molecules Review Role of GSH and Iron-Sulfur Glutaredoxins in Iron Metabolism—Review 1, 1, 1 1 Trnka Daniel y , Hossain Md Faruq y , Jordt Laura Magdalena , Gellert Manuela and Lillig Christopher Horst 2,* 1 Institute for Medical Biochemistry and Molecular Biology, University Medicine, University of Greifswald, 17475 Greifswald, Germany; [email protected] (T.D.); [email protected] (H.M.F.); [email protected] (J.L.M.); [email protected] (G.M.) 2 Christopher Horst Lillig, Institute for Medical Biochemistry and Molecular Biology, University Medicine Greifswald, Ferdinand-Sauerbruch-Straße, 17475 Greifswald, Germany * Correspondence: [email protected]; Tel.: +49-3834-865407; Fax: +49-3834-865402 These authors contributed equally to this work. y Academic Editor: Pál Perjési Received: 29 July 2020; Accepted: 22 August 2020; Published: 25 August 2020 Abstract: Glutathione (GSH) was initially identified and characterized for its redox properties and later for its contributions to detoxification reactions. Over the past decade, however, the essential contributions of glutathione to cellular iron metabolism have come more and more into focus. GSH is indispensable in mitochondrial iron-sulfur (FeS) cluster biosynthesis, primarily by co-ligating FeS clusters as a cofactor of the CGFS-type (class II) glutaredoxins (Grxs). GSH is required for the export of the yet to be defined FeS precursor from the mitochondria to the cytosol. In the cytosol, it is an essential cofactor, again of the multi-domain CGFS-type Grxs, master players in cellular iron and FeS trafficking. In this review, we summarize the recent advances and progress in this field. The most urgent open questions are discussed, such as the role of GSH in the export of FeS precursors from mitochondria, the physiological roles of the CGFS-type Grx interactions with BolA-like proteins and the cluster transfer between Grxs and recipient proteins. -

Difluorodeoxyguanosine in Chinese Hamster Ovary Cells'
[CANCER RESEARCH55, 1517-1524, April 1, 19951 Cytotoxicity, Metabolism, and Mechanisms of Action of 2',2'- Difluorodeoxyguanosine in Chinese Hamster Ovary Cells' Varsha Gandhi,2 Shin Mineishi, Peng Huang, Amy J. Chapman, Yandan Yang, Feng Chen, Biffie Nowak, Shern Chubb, Larry W. Hertel, and William Plunkett Department of Clinical Investigation, The University of Texas M. D. Anderson Cancer Center, Houston, Texas 77030 (V. G., S. M., P. H.. A. J. C.. Y. 1'., F. C., B. N., S. C., w. p.]. andLillyResearchLaboratories,Indianapolis,indiana46285(L W.H.) ABSTRACT treatment of hematological malignancies (4—6).Arabinosyladenine, although not successful in cancer therapy, has proven antiviral activity The emerging clinical success of gemcItabine (2',2'-difluorodeoxycytl dine) Stimulated interest in the synthesis and evaluation of purine conge (7). The importance of the 2'-pentose moiety for the design of chemo ners. The cytotoxicity, metabolism, and mechanisms of action of the lead therapeutic agents and the fact that fluorine has a van der Waals radius candidate, 2',2'-difluorodeoxyguanoslne (dFdGuo), were StUdied In ChI nese hamster ovary cells Unlike the natural nucleoside deoxyguanosine similar to that of hydrogen led to the synthesis and evaluation of (dGuo), dFdGuo was not a substrate for purine nucleoside phosphorylase 2'-fluorinated analogues. 2'-Deoxy-2'-fluorocytidine and 2'-deoxy Wild-type Chinese hamster ovary cells and a mutant line deficient in 2'-fluoroguanosine possess inhibitory activities against several deoxycytidine (dCyd) kinase were similarly affected by dFdGuo (50% lymphoid cell lines and influenza viruses, respectively (8, 9). Inter inhibitory concentration, 7.5 and 6.5 pM, respectively), suggesting that estingly, substitution of the 2'-hydrogen of dCyd3 with a fluorine unlike gemcitabine, dCyd kinase was not responsible for activation of atom in the arabinose configuration produced 10-fold greater cyto dFdGuo This was further confirmed by separation of nucleoside kinases toxicity than when placed in the ribose position (10). -

DHFR Inhibitors: Reading the Past for Discovering Novel Anticancer Agents
molecules Review DHFR Inhibitors: Reading the Past for Discovering Novel Anticancer Agents Maria Valeria Raimondi 1,*,† , Ornella Randazzo 1,†, Mery La Franca 1 , Giampaolo Barone 1 , Elisa Vignoni 2, Daniela Rossi 2 and Simona Collina 2,* 1 Department of Biological, Chemical and Pharmaceutical Sciences and Technologies (STEBICEF), University of Palermo, via Archirafi 32, 90123 Palermo, Italy; [email protected] (O.R.); [email protected] (M.L.F.); [email protected] (G.B.) 2 Drug Sciences Department, Medicinal Chemistry and Pharmaceutical Technology Section, University of Pavia, via Taramelli 12, 27100 Pavia, Italy; [email protected] (E.V.); [email protected] (D.R.) * Correspondence: [email protected] (M.V.R.); [email protected] (S.C.); Tel.: +390-912-389-1915 (M.V.R.); +390-382-987-379 (S.C.) † These Authors contributed equally to this work. Academic Editors: Simona Collina and Mariarosaria Miloso Received: 25 February 2019; Accepted: 20 March 2019; Published: 22 March 2019 Abstract: Dihydrofolate reductase inhibitors are an important class of drugs, as evidenced by their use as antibacterial, antimalarial, antifungal, and anticancer agents. Progress in understanding the biochemical basis of mechanisms responsible for enzyme selectivity and antiproliferative effects has renewed the interest in antifolates for cancer chemotherapy and prompted the medicinal chemistry community to develop novel and selective human DHFR inhibitors, thus leading to a new generation of DHFR inhibitors. This work summarizes the mechanism of action, chemical, and anticancer profile of the DHFR inhibitors discovered in the last six years. New strategies in DHFR drug discovery are also provided, in order to thoroughly delineate the current landscape for medicinal chemists interested in furthering this study in the anticancer field. -

Biochemical and Biophysical Characterization of a Mycoredoxin
International Journal of Biological Macromolecules 107 (2018) 1999–2007 Contents lists available at ScienceDirect International Journal of Biological Macromolecules j ournal homepage: www.elsevier.com/locate/ijbiomac Biochemical and biophysical characterization of a mycoredoxin protein glutaredoxin A1 from Corynebacterium pseudotuberculosis a a a b a Raphael J. Eberle , Liege A. Kawai , Fabio R. de Moraes , Ljubica Tasic , Raghuvir K. Arni , a,∗ Monika A. Coronado a Multiuser Center for Biomolecular Innovation, Departament of Physics, Instituto de Biociências Letras e Ciências Exatas (Ibilce), Universidade Estadual Paulista (UNESP), São Jose do Rio Preto, SP, 15054-000, Brazil b Institute of Chemistry, University of Campinas (UNICAMP), Campinas, SP, 13083-970, Brazil a r t i c l e i n f o a b s t r a c t Article history: Glutaredoxin A1 from Corynebacterium pseudotuberculosis was shown to be a mycoredoxin protein. In Received 25 July 2017 this study, we established a process to overexpress and purify glutaredoxin A1. The aim of this study Received in revised form 6 October 2017 was the investigation of the Glutaredoxin A1 from C. pseudotuberculosis behavior under different redox Accepted 11 October 2017 environments and the identification of lead molecules, which can be used for specific inhibitor develop- Available online 16 October 2017 ment for this protein family. A quantitative assay was performed measuring the rate of insulin reduction spectrophotometrically at 640 nm through turbidity formation from the precipitation of the free insulin. Keywords: Glutaredoxin A1, at 5 M concentration, accelerated the reduction process of 0.2 mM insulin and 1 mM Corynebacterium pseudotuberculosis Glutaredoxin DTT. -

1611 REGULATION of PYRIMIDINE METABOLISM in PLANTS Chris
[Frontiers in Bioscience 9, 1611-1625, May 1, 2004] REGULATION OF PYRIMIDINE METABOLISM IN PLANTS 1, 2 1, 3 1, 4 1, 5 1, 6 1, 7 Chris Kafer , Lan Zhou , Djoko Santoso , Adel Guirgis , Brock Weers , Sanggyu Park and Robert Thornburg 1 1 Department of Biochemistry, Biophysics, and Molecular Biology, Iowa State University, Ames, Iowa 50011, 2 BASF Plant Science LLC, 2901 South Loop Drive, Ste 3800, Ames, Iowa 50014, 3 Lan Zhou, Pioneer Hi-Bred International, Inc. 7300 NW 62nd Avenue, PO Box 1004, Johnston, Iowa 50131-1004, 4 Indonesian Biotechnology Research Institute for Estate Crops, Jl, Taman Kencana No 1, Bogor 16151 Indonesia, 5 Institute of Genetic Engineering and Biotechnology, Menofiya University, PO Box 79/22857, Sadat City, Egypt, 6 Department of Biochemistry, University of Iowa, 4/511 Bowen Science Building, Iowa City, Iowa 52242-1109, 7 Division of Life and Environment, College of Natural Resources, Daegu University, Gyongsan City, Gyongbuk, Korea 712-714 TABLE OF CONTENTS 1. Abstract 2. Introduction 3. Pyrimidine metabolic pathways 3.1. De novo pyrimidine biosynthesis 3.1.1. CPSase 3.1.2. ATCase 3.1.3. DHOase 3.1.4. DHODH 3.1.5. UMPS 3.1.6. Intracellular Organization of the de novo Pathway 3.2. Pyrimidine Salvage and Recycling 3.2.1. Cytosine deaminase 3.2.2. Cytidine deaminase 3.2.3. UPRTase 3.3. Pyrimidine Modification 3.3.1. UMP/CMP kinase 3.3.2. NDP kinase 3.3.3. CTP synthase, NDP reductase, dUTPase 3.3.4. Thymidylate synthase/Dihydrofolate reductase 3.4. Pyrimidine Catabolism 4. Regulation of pyrimidine metabolism 4.1.