A Proteomic Approach for Studying Insect Phylogeny: CAPA Peptides of Ancient Insect Taxa (Dictyoptera, Blattoptera) As a Test Case
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

New Species of Hammerschmidtiella Chitwood, 1932, and Blattophila
Zootaxa 4226 (3): 429–441 ISSN 1175-5326 (print edition) http://www.mapress.com/j/zt/ Article ZOOTAXA Copyright © 2017 Magnolia Press ISSN 1175-5334 (online edition) https://doi.org/10.11646/zootaxa.4226.3.6 http://zoobank.org/urn:lsid:zoobank.org:pub:77877607-ECE7-455E-A76C-353B16F92296 New species of Hammerschmidtiella Chitwood, 1932, and Blattophila Cobb, 1920, and new geographical records for Severianoia annamensis Van Luc & Spiridonov, 1993 (Nematoda: Oxyurida: Thelastomatoidea) from Cockroaches (Insecta: Blattaria) in Ohio and Florida, U.S.A. RAMON A. CARRENO Department of Zoology, Ohio Wesleyan University, Delaware, Ohio, 43015, USA. E-mail: [email protected] Abstract Two new species of thelastomatid nematodes parasitic in the hindgut of cockroaches are described. Hammerschmidtiella keeneyi n. sp. is described from a laboratory colony of Diploptera punctata (Eschscholtz, 1822) from a facility in Ohio, U. S. A. This species is characterized by having females with a short tail and males smaller than those described from other species. The new species also differs from others in the genus by a number of differing measurements that indicate a distinct identity, including esophageal, tail, and egg lengths as well as the relative position of the excretory pore. Blat- tophila peregrinata n. sp. is described from Periplaneta australasiae (Fabricius, 1775) and Pycnoscelus surinamensis (Linnaeus, 1758) in a greenhouse from Ohio, U.S.A. and from wild P. surinamensis in southern Florida, U.S.A. This spe- cies differs from others in the genus by having a posteriorly directed vagina, vulva in the anterior third of the body, no lateral alae in females, and eggs with an operculum. -

Isoptera Book Chapter
Isoptera 535 See Also the Following Articles Biodiversity ■ Biogeographical Patterns ■ Cave Insects ■ Introduced Insects Further Reading Carlquist , S. ( 1974 ) . “ Island Biology . ” Columbia University Press , New York and London . Gillespie , R. G. , and Roderick , G. K. ( 2002 ) . Arthropods on islands: Colonization, speciation, and conservation . Annu. Rev. Entomol. 47 , 595 – 632 . Gillespie , R. G. , and Clague , D. A. (eds.) (2009 ) . “ Encyclopedia of Islands. ” University of California Press , Berkeley, CA . Howarth , F. G. , and Mull , W. P. ( 1992 ) . “ Hawaiian Insects and Their Kin . ” University of Hawaii Press , Honolulu, HI . MacArthur , R. H. , and Wilson , E. O. ( 1967 ) . “ The Theory of Island Biogeography . ” Princeton University Press , Princeton, NJ . Wagner , W. L. , and Funk , V. (eds.) ( 1995 ) . “ Hawaiian Biogeography Evolution on a Hot Spot Archipelago. ” Smithsonian Institution Press , Washington, DC . Whittaker , R. J. , and Fern á ndez-Palacios , J. M. ( 2007 ) . “ Island Biogeography: Ecology, Evolution, and Conservation , ” 2nd ed. Oxford University Press , Oxford, U.K . I Isoptera (Termites) Vernard R. Lewis FIGURE 1 Castes for Isoptera. A lower termite group, University of California, Berkeley Reticulitermes, is represented. A large queen is depicted in the center. A king is to the left of the queen. A worker and soldier are he ordinal name Isoptera is of Greek origin and refers to below. (Adapted, with permission from Aventis Environmental the two pairs of straight and very similar wings that termites Science, from The Mallis Handbook of Pest Control, 1997.) Thave as reproductive adults. Termites are small and white to tan or sometimes black. They are sometimes called “ white ants ” and can be confused with true ants (Hymenoptera). -

Genetically Modified Baculoviruses for Pest
INSECT CONTROL BIOLOGICAL AND SYNTHETIC AGENTS This page intentionally left blank INSECT CONTROL BIOLOGICAL AND SYNTHETIC AGENTS EDITED BY LAWRENCE I. GILBERT SARJEET S. GILL Amsterdam • Boston • Heidelberg • London • New York • Oxford Paris • San Diego • San Francisco • Singapore • Sydney • Tokyo Academic Press is an imprint of Elsevier Academic Press, 32 Jamestown Road, London, NW1 7BU, UK 30 Corporate Drive, Suite 400, Burlington, MA 01803, USA 525 B Street, Suite 1800, San Diego, CA 92101-4495, USA ª 2010 Elsevier B.V. All rights reserved The chapters first appeared in Comprehensive Molecular Insect Science, edited by Lawrence I. Gilbert, Kostas Iatrou, and Sarjeet S. Gill (Elsevier, B.V. 2005). All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopy, recording, or any information storage and retrieval system, without permission in writing from the publishers. Permissions may be sought directly from Elsevier’s Rights Department in Oxford, UK: phone (þ44) 1865 843830, fax (þ44) 1865 853333, e-mail [email protected]. Requests may also be completed on-line via the homepage (http://www.elsevier.com/locate/permissions). Library of Congress Cataloging-in-Publication Data Insect control : biological and synthetic agents / editors-in-chief: Lawrence I. Gilbert, Sarjeet S. Gill. – 1st ed. p. cm. Includes bibliographical references and index. ISBN 978-0-12-381449-4 (alk. paper) 1. Insect pests–Control. 2. Insecticides. I. Gilbert, Lawrence I. (Lawrence Irwin), 1929- II. Gill, Sarjeet S. SB931.I42 2010 632’.7–dc22 2010010547 A catalogue record for this book is available from the British Library ISBN 978-0-12-381449-4 Cover Images: (Top Left) Important pest insect targeted by neonicotinoid insecticides: Sweet-potato whitefly, Bemisia tabaci; (Top Right) Control (bottom) and tebufenozide intoxicated by ingestion (top) larvae of the white tussock moth, from Chapter 4; (Bottom) Mode of action of Cry1A toxins, from Addendum A7. -

Blattabacterium Genome: Structure, Function, and Evolution Austin Alleman
University of Texas at Tyler Scholar Works at UT Tyler Biology Theses Biology Spring 6-5-2014 Blattabacterium Genome: Structure, Function, and Evolution Austin Alleman Follow this and additional works at: https://scholarworks.uttyler.edu/biology_grad Part of the Biology Commons Recommended Citation Alleman, Austin, "Blattabacterium Genome: Structure, Function, and Evolution" (2014). Biology Theses. Paper 20. http://hdl.handle.net/10950/215 This Thesis is brought to you for free and open access by the Biology at Scholar Works at UT Tyler. It has been accepted for inclusion in Biology Theses by an authorized administrator of Scholar Works at UT Tyler. For more information, please contact [email protected]. BLATTABACTERIUM GENOME: STRUCTURE, FUNCTION, AND EVOLUTION by AUSTIN ALLEMAN A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science Department of Biology Srini Kambhampati, Ph. D., Committee Chair College of Arts and Sciences The University of Texas at Tyler May 2014 © Copyright by Austin Alleman 2014 All rights reserved Acknowledgements I would like to thank the Sam A. Lindsey Endowment, without whose support this research would not have been possible. I would also like to thank my research advisor, Dr. Srini Kambhampati, for his knowledge, insight, and patience; and my committee, Dr. John S. Placyk, Jr. and Dr. Blake Bextine, for their assistance and feedback. “It is the supreme art of the teacher to awaken joy in creative expression and knowledge.” --Albert Einstein Table of Contents Chapter -

Bulletin Number / Numéro 2 Entomological Society of Canada Société D’Entomologie Du Canada June / Juin 2008
Volume 40 Bulletin Number / numéro 2 Entomological Society of Canada Société d’entomologie du Canada June / juin 2008 Published quarterly by the Entomological Society of Canada Publication trimestrielle par la Société d’entomologie du Canada ............................................................... .................................................................................................................................................................................................................................................................................................................................. .......................................................................... ........................................................................................................................................................................ ....................... ................................................................................. ................................................. List of contents / Table des matières Volume 40 (2), June / june 2008 Up front / Avant-propos ................................................................................................................49 Moth balls / Boules à mites .............................................................................................................51 Meeting announcements / Réunions futures ..................................................................................52 Dear Buggy / Cher Bibitte ..............................................................................................................53 -

Moniliformis Moniliformis Infection Has No Effect on Some Behaviors of the Cockroach Diploptera Punctata Author(S): Zachary Allely, Janice Moore and Nicholas J
Moniliformis moniliformis Infection Has No Effect on Some Behaviors of the Cockroach Diploptera punctata Author(s): Zachary Allely, Janice Moore and Nicholas J. Gotelli Source: The Journal of Parasitology, Vol. 78, No. 3 (Jun., 1992), pp. 524-526 Published by: The American Society of Parasitologists Stable URL: http://www.jstor.org/stable/3283658 . Accessed: 04/05/2013 10:55 Your use of the JSTOR archive indicates your acceptance of the Terms & Conditions of Use, available at . http://www.jstor.org/page/info/about/policies/terms.jsp . JSTOR is a not-for-profit service that helps scholars, researchers, and students discover, use, and build upon a wide range of content in a trusted digital archive. We use information technology and tools to increase productivity and facilitate new forms of scholarship. For more information about JSTOR, please contact [email protected]. The American Society of Parasitologists is collaborating with JSTOR to digitize, preserve and extend access to The Journal of Parasitology. http://www.jstor.org This content downloaded from 132.198.40.214 on Sat, 4 May 2013 10:55:35 AM All use subject to JSTOR Terms and Conditions RESEARCH NOTES J. Parasitol., 78(3), 1992, p. 524-526 ? American Society of Parasitologists 1992 Moniliformismoniliformis Infection Has No Effect on Some Behaviorsof the CockroachDiploptera punctata Zachary Allely, Janice Moore, and Nicholas J. Gotelli*, Departmentof Biology,Colorado State University,Fort Collins,Colorado 80523; and *Departmentof Zoology,University of Oklahoma,Norman, Oklahoma 73019 ABSTRACT: The behaviorof the cockroachDiploptera Experiments were conducted under 2 light punctataparasitized with the acanthocephalanMonil- conditions: white light and red light (Carmichael was examined for iformis moniliformis parasite-in- and Moore, 1991). -
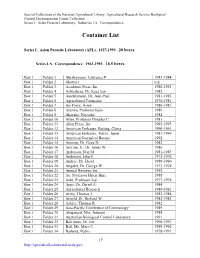
Container List
Special Collections of the National Agricultural Library: Agricultural Research Service Biological Control Documentation Center Collection Series I. Asian Parasite Laboratory. Subseries I.A. Correspondence. Container List Series I. Asian Parasite Laboratory (APL). 1927-1993. 20 boxes. Series I.A. Correspondence. 1963-1993. 16.5 boxes. Box 1 Folder 1 Abrahamson, Lawrence P. 1983-1984 Box 1 Folder 2 Abstract n.d. Box 1 Folder 3 Academic Press, Inc. 1986-1993 Box 1 Folder 4 Achterberg, Dr. Kees van 1982 Box 1 Folder 5 Aeschlimann, Dr. Jean-Paul 1981-1985 Box 1 Folder 6 Agricultural Counselor 1976-1981 Box 1 Folder 7 Air Force, Army 1980-1981 Box 1 Folder 8 Aizawa, Professor Keio 1986 Box 1 Folder 9 Akasaka, Naoyuki 1982 Box 1 Folder 10 Allen, Professor Douglas C. 1981 Box 1 Folder 11 Allen Press, Inc. 1992-1993 Box 1 Folder 12 American Embassy, Beijing, China 1990-1991 Box 1 Folder 13 American Embassy, Tokyo, Japan 1981-1990 Box 1 Folder 14 American Journal of Botany 1992 Box 1 Folder 15 Amman, Dr. Gene D. 1982 Box 1 Folder 16 Amrine, Jr., Dr. James W. 1986 Box 1 Folder 17 Anderson, Don M. 1981-1983 Box 1 Folder 18 Anderson, John F. 1975-1976 Box 1 Folder 19 Andres, Dr. Lloyd 1989-1990 Box 1 Folder 20 Angalet, Dr. George W. 1973-1978 Box 1 Folder 21 Annual Reviews Inc. 1992 Box 1 Folder 22 Ao, Professor Hsien-Bine 1988 Box 1 Folder 23 Aoki, Professor Joji 1977-1978 Box 1 Folder 24 Apps, Dr. Darrel A. 1984 Box 1 Folder 25 Agricultural Research 1980-1981 Box 1 Folder 26 Army, Thomas J. -

The Phylogeny of Termites
Molecular Phylogenetics and Evolution 48 (2008) 615–627 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev The phylogeny of termites (Dictyoptera: Isoptera) based on mitochondrial and nuclear markers: Implications for the evolution of the worker and pseudergate castes, and foraging behaviors Frédéric Legendre a,*, Michael F. Whiting b, Christian Bordereau c, Eliana M. Cancello d, Theodore A. Evans e, Philippe Grandcolas a a Muséum national d’Histoire naturelle, Département Systématique et Évolution, UMR 5202, CNRS, CP 50 (Entomologie), 45 rue Buffon, 75005 Paris, France b Department of Integrative Biology, 693 Widtsoe Building, Brigham Young University, Provo, UT 84602, USA c UMR 5548, Développement—Communication chimique, Université de Bourgogne, 6, Bd Gabriel 21000 Dijon, France d Muzeu de Zoologia da Universidade de São Paulo, Avenida Nazaré 481, 04263-000 São Paulo, SP, Brazil e CSIRO Entomology, Ecosystem Management: Functional Biodiversity, Canberra, Australia article info abstract Article history: A phylogenetic hypothesis of termite relationships was inferred from DNA sequence data. Seven gene Received 31 October 2007 fragments (12S rDNA, 16S rDNA, 18S rDNA, 28S rDNA, cytochrome oxidase I, cytochrome oxidase II Revised 25 March 2008 and cytochrome b) were sequenced for 40 termite exemplars, representing all termite families and 14 Accepted 9 April 2008 outgroups. Termites were found to be monophyletic with Mastotermes darwiniensis (Mastotermitidae) Available online 27 May 2008 as sister group to the remainder of the termites. In this remainder, the family Kalotermitidae was sister group to other families. The families Kalotermitidae, Hodotermitidae and Termitidae were retrieved as Keywords: monophyletic whereas the Termopsidae and Rhinotermitidae appeared paraphyletic. -

Running Speed and Food Intake of the Matrotrophic Viviparous Cockroach Diploptera Punctata (Blattodea: Blaberidae) During Gestation
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Entomologie heute Jahr/Year: 2014 Band/Volume: 26 Autor(en)/Author(s): Greven Hartmut, Floßdorf David, Köthe Janine, List Fabian, Zwanzig Nadine Artikel/Article: Running Speed and Food Intake of the Matrotrophic Viviparous Cockroach Diploptera punctata (Blattodea: Blaberidae) during Gestation. Laufgeschwindigkeit und Nahrungsaufnahme der matrotroph viviparen Schabe Diploptera punctata (Blattodea: Blaberidae) während der Trächtigkeit 53-72 Running speed and food intake of Diploptera punctata during gestation 53 Entomologie heute 26 (2014): 53-72 Running Speed and Food Intake of the Matrotrophic Viviparous Cockroach Diploptera punctata (Blattodea: Blaberidae) during Gestation Laufgeschwindigkeit und Nahrungsaufnahme der matrotroph viviparen Schabe Diploptera punctata (Blattodea: Blaberidae) während der Trächtigkeit HARTMUT GREVEN, DAVID FLOSSDORF, JANINE KÖTHE, FABIAN LIST & NADINE ZWANZIG Summary: Diploptera punctata is the only cockroach, which has been clearly characterized as matro- trophic viviparous. Our observations on courtship and mating generally confi rm the data from the literature. Courtship and mating correspond to type I (male offers himself under wing fl uttering, female mounts the male, nibbles on his tergal glands, dismounts, turns to achieve the fi nal mating position, i.e. abdomen to abdomen, heads in the opposite direction. We document photographic ally mating and courtship of fully sklerotized, sexually experienced males with teneral females imme- diately after the last moult, and with fully sclerotized females several hours after the fi nal moult. Effects of sexual dimorphism (females are larger than males) and pregnancy (females gain weight) became apparent from the running speed cockroaches reached, when disturbed. Males and females ran signifi cantly faster during daytime than at night, but males ran always faster than females. -

Long Rdna Amplicon Sequencing of Insect-Infecting Nephridiophagids
www.nature.com/scientificreports OPEN Long rDNA amplicon sequencing of insect‑infecting nephridiophagids reveals their afliation to the Chytridiomycota and a potential to switch between hosts Jürgen F. H. Strassert 1*, Christian Wurzbacher 2, Vincent Hervé 3, Taraha Antany1, Andreas Brune 3 & Renate Radek 1* Nephridiophagids are unicellular eukaryotes that parasitize the Malpighian tubules of numerous insects. Their life cycle comprises multinucleate vegetative plasmodia that divide into oligonucleate and uninucleate cells, and sporogonial plasmodia that form uninucleate spores. Nephridiophagids are poor in morphological characteristics, and although they have been tentatively identifed as early‑branching fungi based on the SSU rRNA gene sequences of three species, their exact position within the fungal tree of live remained unclear. In this study, we describe two new species of nephridiophagids (Nephridiophaga postici and Nephridiophaga javanicae) from cockroaches. Using long‑read sequencing of the nearly complete rDNA operon of numerous further species obtained from cockroaches and earwigs to improve the resolution of the phylogenetic analysis, we found a robust afliation of nephridiophagids with the Chytridiomycota—a group of zoosporic fungi that comprises parasites of diverse host taxa, such as microphytes, plants, and amphibians. The presence of the same nephridiophagid species in two only distantly related cockroaches indicates that their host specifcity is not as strict as generally assumed. Insects are the most diverse group of all animals. So far, about one million species have been described and recent estimates for extant species range from 2.6 to 7.8 million1,2. Tey are globally distributed and impact human life at numerous levels. In agriculture, for instance, insects play a major role as both pollinators (e.g., honey bees) and pests that feed on crops (e.g., grasshoppers). -

Univerzita Palackého V Olomouci Pedagogická
UNIVERZITA PALACKÉHO V OLOMOUCI PEDAGOGICKÁ FAKULTA KATEDRA BIOLOGIE Bc. Tereza Friedlová Učitelství přírodopisu a matematiky pro 2. st. ZŠ Rodová revize světluškovitých brouků podčeledi Amydetinae (Coleoptera: Lampyridae) Diplomová práce Vedoucí diplomové práce: Prof.Ing. Milada Bocáková, Ph.D. OLOMOUC 2015 Prohlašuji, že jsem tuto diplomovou práci napsala samostatně pod vedením Prof. Ing. Milady Bocákové, Ph.D. s využitím uvedené literatury. V Olomouci dne 17. 6. 2015 …………………………………………. Bc. Tereza Friedlová Děkuji vedoucí diplomové práce Prof. Ing. Miladě Bocákové, Ph.D za vedení diplomové práce, cenné rady a pomoc při získávání potřebných materiálů. OBSAH ÚVOD ................................................................................................................................... 5 1 BIOLUMINISCENCE .................................................................................................... 9 1.1 Bioluminiscence světlušek ........................................................................................ 10 1.1.1 Luminiscence larev .............................................................................................. 12 1.1.2 Luminiscence dospělých ...................................................................................... 13 1.2 Komunikační signály ................................................................................................ 14 1.3 Přehled názorů na evoluci bioluminiscence ............................................................... 16 2 HISTORIE ČELEDI LAMPYRIDAE SE ZAMĚŘENÍM -

Towards Classical Biological Control of Leek Moth
____________________________________________________________________________ Ateyyat This project seeks to provide greater coherence for the biocontrol knowledge system for regulators and researchers; create an open access information source for biocontrol re- search of agricultural pests in California, which will stimulate greater international knowl- edge sharing about agricultural pests in Mediterranean climates; and facilitate the exchange of information through a cyberinfrastructure among government regulators, and biocontrol entomologists and practitioners. It seeks broader impacts through: the uploading of previ- ously unavailable data being made openly accessible; the stimulation of greater interaction between the biological control regulation, research, and practitioner community in selected Mediterranean regions; the provision of more coherent and useful information to enhance regulatory decisions by public agency scientists; a partnership with the IOBC to facilitate international data sharing; and progress toward the ultimate goal of increasing the viability of biocontrol as a reduced risk pest control strategy. No Designated Session Theme BIOLOGY OF CIRROSPILUS INGENUUS GAHAN (HYMENOPTERA: EULOPHIDAE), AN ECTOPARASITOID OF THE CITRUS LEAFMINER, PHYLLOCNISTIS CITRELLA STAINTON (LEPIDOPTERA: GRACILLARIIDAE) ON LEMON 99 Mazen A. ATEYYAT Al-Shoubak University College, Al-Balqa’ Applied University, P.O. Box (5), Postal code 71911, Al-Shawbak, Jordan [email protected] The citrus leafminer (CLM), Phyllocnistis citrella Stainton (Lepidoptera: Gracillariidae) in- vaded the Jordan Valley in 1994 and was able to spread throughout Jordan within a few months of its arrival. It was the most common parasitoid from 1997 to 1999 in the Jordan Valley. An increase in the activity of C. ingenuus was observed in autumn and the highest number of emerged C. ingenuus adults was in November 1999.