Chapter 2 Introduction to Some Basic Features of Genetic Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Glossary - Cellbiology
1 Glossary - Cellbiology Blotting: (Blot Analysis) Widely used biochemical technique for detecting the presence of specific macromolecules (proteins, mRNAs, or DNA sequences) in a mixture. A sample first is separated on an agarose or polyacrylamide gel usually under denaturing conditions; the separated components are transferred (blotting) to a nitrocellulose sheet, which is exposed to a radiolabeled molecule that specifically binds to the macromolecule of interest, and then subjected to autoradiography. Northern B.: mRNAs are detected with a complementary DNA; Southern B.: DNA restriction fragments are detected with complementary nucleotide sequences; Western B.: Proteins are detected by specific antibodies. Cell: The fundamental unit of living organisms. Cells are bounded by a lipid-containing plasma membrane, containing the central nucleus, and the cytoplasm. Cells are generally capable of independent reproduction. More complex cells like Eukaryotes have various compartments (organelles) where special tasks essential for the survival of the cell take place. Cytoplasm: Viscous contents of a cell that are contained within the plasma membrane but, in eukaryotic cells, outside the nucleus. The part of the cytoplasm not contained in any organelle is called the Cytosol. Cytoskeleton: (Gk. ) Three dimensional network of fibrous elements, allowing precisely regulated movements of cell parts, transport organelles, and help to maintain a cell’s shape. • Actin filament: (Microfilaments) Ubiquitous eukaryotic cytoskeletal proteins (one end is attached to the cell-cortex) of two “twisted“ actin monomers; are important in the structural support and movement of cells. Each actin filament (F-actin) consists of two strands of globular subunits (G-Actin) wrapped around each other to form a polarized unit (high ionic cytoplasm lead to the formation of AF, whereas low ion-concentration disassembles AF). -

Comparison of the Effects on Mrna and Mirna Stability Arian Aryani and Bernd Denecke*
Aryani and Denecke BMC Research Notes (2015) 8:164 DOI 10.1186/s13104-015-1114-z RESEARCH ARTICLE Open Access In vitro application of ribonucleases: comparison of the effects on mRNA and miRNA stability Arian Aryani and Bernd Denecke* Abstract Background: MicroRNA has become important in a wide range of research interests. Due to the increasing number of known microRNAs, these molecules are likely to be increasingly seen as a new class of biomarkers. This is driven by the fact that microRNAs are relatively stable when circulating in the plasma. Despite extensive analysis of mechanisms involved in microRNA processing, relatively little is known about the in vitro decay of microRNAs under defined conditions or about the relative stabilities of mRNAs and microRNAs. Methods: In this in vitro study, equal amounts of total RNA of identical RNA pools were treated with different ribonucleases under defined conditions. Degradation of total RNA was assessed using microfluidic analysis mainly based on ribosomal RNA. To evaluate the influence of the specific RNases on the different classes of RNA (ribosomal RNA, mRNA, miRNA) ribosomal RNA as well as a pattern of specific mRNAs and miRNAs was quantified using RT-qPCR assays. By comparison to the untreated control sample the ribonuclease-specific degradation grade depending on the RNA class was determined. Results: In the present in vitro study we have investigated the stabilities of mRNA and microRNA with respect to the influence of ribonucleases used in laboratory practice. Total RNA was treated with specific ribonucleases and the decay of different kinds of RNA was analysed by RT-qPCR and miniaturized gel electrophoresis. -

Exploring the Structure of Long Non-Coding Rnas, J
IMF YJMBI-63988; No. of pages: 15; 4C: 3, 4, 7, 8, 10 1 2 Rise of the RNA Machines: Exploring the Structure of 3 Long Non-Coding RNAs 4 Irina V. Novikova, Scott P. Hennelly, Chang-Shung Tung and Karissa Y. Sanbonmatsu Q15 6 Los Alamos National Laboratory, Los Alamos, NM 87545, USA 7 Correspondence to Karissa Y. Sanbonmatsu: [email protected] 8 http://dx.doi.org/10.1016/j.jmb.2013.02.030 9 Edited by A. Pyle 1011 12 Abstract 13 Novel, profound and unexpected roles of long non-coding RNAs (lncRNAs) are emerging in critical aspects of 14 gene regulation. Thousands of lncRNAs have been recently discovered in a wide range of mammalian 15 systems, related to development, epigenetics, cancer, brain function and hereditary disease. The structural 16 biology of these lncRNAs presents a brave new RNA world, which may contain a diverse zoo of new 17 architectures and mechanisms. While structural studies of lncRNAs are in their infancy, we describe existing 18 structural data for lncRNAs, as well as crystallographic studies of other RNA machines and their implications 19 for lncRNAs. We also discuss the importance of dynamics in RNA machine mechanism. Determining 20 commonalities between lncRNA systems will help elucidate the evolution and mechanistic role of lncRNAs in 21 disease, creating a structural framework necessary to pursue lncRNA-based therapeutics. 22 © 2013 Published by Elsevier Ltd. 24 23 25 Introduction rather than the exception in the case of eukaryotic 50 organisms. 51 26 RNA is primarily known as an intermediary in gene LncRNAs are defined by the following: (i) lack of 52 11 27 expression between DNA and proteins. -

RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases
G C A T T A C G G C A T genes Review RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases Amber Willbanks, Shaun Wood and Jason X. Cheng * Department of Pathology, Hematopathology Section, University of Chicago, Chicago, IL 60637, USA; [email protected] (A.W.); [email protected] (S.W.) * Correspondence: [email protected] Abstract: Chromatin structure plays an essential role in eukaryotic gene expression and cell identity. Traditionally, DNA and histone modifications have been the focus of chromatin regulation; however, recent molecular and imaging studies have revealed an intimate connection between RNA epigenetics and chromatin structure. Accumulating evidence suggests that RNA serves as the interplay between chromatin and the transcription and splicing machineries within the cell. Additionally, epigenetic modifications of nascent RNAs fine-tune these interactions to regulate gene expression at the co- and post-transcriptional levels in normal cell development and human diseases. This review will provide an overview of recent advances in the emerging field of RNA epigenetics, specifically the role of RNA modifications and RNA modifying proteins in chromatin remodeling, transcription activation and RNA processing, as well as translational implications in human diseases. Keywords: 5’ cap (5’ cap); 7-methylguanosine (m7G); R-loops; N6-methyladenosine (m6A); RNA editing; A-to-I; C-to-U; 2’-O-methylation (Nm); 5-methylcytosine (m5C); NOL1/NOP2/sun domain Citation: Willbanks, A.; Wood, S.; (NSUN); MYC Cheng, J.X. RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases. Genes 2021, 12, 627. -

SARS-Cov-2 RNA, Qualitative Real-Time RT-PCR (Test Code 39433)
SARS-CoV-2 RNA, Qualitative Real-Time RT-PCR (Test Code 39433) Package Insert For Emergency Use Only For In-vitro Diagnostic Use - Rx Only Intended Use The Quest Diagnostics SARS-CoV-2 RNA, Qualitative Real-Time RT-PCR (“Quest SARS-CoV-2 rRT-PCR”) is a real-time RT-PCR test intended for the qualitative detection of nucleic acid from the SARS-CoV-2 in upper and lower respiratory specimens (such as nasopharyngeal or oropharyngeal swabs, sputum, tracheal aspirates, and bronchoalveolar lavage) collected from individuals suspected of COVID-19 by their healthcare provider. This test is also for use with nasal swab specimens that are self-collected at home or in a healthcare setting by individuals using an authorized home-collection kit when determined to be appropriate by a healthcare provider. This test is for the qualitative detection of nucleic acid from the SARS-CoV-2 in pooled samples containing up to four of the individual upper respiratory swab specimens (nasopharyngeal, mid-turbinate, anterior nares or oropharyngeal swabs) that were collected in individual vials containing transport media from individuals suspected of COVID-19 by their healthcare provider. Negative results from pooled testing should not be treated as definitive. If patient’s clinical signs and symptoms are inconsistent with a negative result or results are necessary for patient management, then the patient should be considered for individual testing. Specimens included in pools with a positive, inconclusive, or invalid result must be tested individually prior to reporting a result. Specimens with low viral loads may not be detected in sample pools due to the decreased sensitivity of pooled testing. -

The Difficult Case of an RNA-Only Origin of Life
Emerging Topics in Life Sciences (2019) 3 469–475 https://doi.org/10.1042/ETLS20190024 Perspective The difficult case of an RNA-only origin of life Kristian Le Vay and Hannes Mutschler Max-Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany Downloaded from https://portlandpress.com/emergtoplifesci/article-pdf/3/5/469/859756/etls-2019-0024c.pdf by Max-Planck-Institut fur Biochemie user on 28 November 2019 Correspondence: Hannes Mutschler ([email protected]) The RNA world hypothesis is probably the most extensively studied model for the emergence of life on Earth. Despite a large body of evidence supporting the idea that RNA is capable of kick-starting autocatalytic self-replication and thus initiating the emergence of life, seemingly insurmountable weaknesses in the theory have also been highlighted. These problems could be overcome by novel experimental approaches, including out-of- equilibrium environments, and the exploration of an early co-evolution of RNA and other key biomolecules such as peptides and DNA, which might be necessary to mitigate the shortcomings of RNA-only systems. The conjecture that life on Earth evolved from an ‘RNA World’ remains one of the most popular hypotheses for abiogenesis, even 60 years after Alex Rich first put the idea forward [1]. For some, evi- dence based upon ubiquitous molecular fossils and the elegance of the idea that RNA once had a dual role as information carrier and prebiotic catalyst provide overwhelming support for the theory. Nevertheless, doubts remain surrounding the chemical evolution of an RNA world, whose classical scenario is based on a temporal sequence of nucleotide formation, enzyme-free polymerisation/replica- tion, recombination, encapsulation in lipid vesicles (or other compartments), evolution of ribozymes and finally the innovation of the genetic code and its translation (Figure 1)[2,3]. -
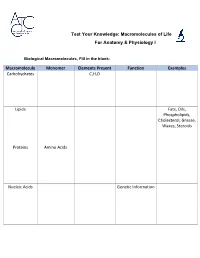
Macromolecules of Life Worksheet
Test Your Knowledge: Macromolecules of Life For Anatomy & Physiology I Biological Macromolecules, Fill in the blank: Macromolecule Monomer Elements Present Function Examples Carbohydrates C,H,O Lipids Fats, Oils, Phospholipids, Cholesterol, Grease, Waxes, Steroids Proteins Amino Acids Nucleic Acids Genetic Information Carbohydrates are classified by __________. The most common simple sugars are glucose, galactose and fructose that are made of a single sugar molecule. These can be classified as __________. Sucrose and __________ are classified as disaccharides; they are made of two monosaccharides joined by a dehydration reaction. The most complex carbohydrates are starch, __________ and cellulose, classified as __________. Lipids most abundant form are __________. Triglycerides building blocks are 1 __________ and 3 __________ per molecule. If a triglyceride only contains __________ bonds that contain the maximum number of __________, then it is classified as a saturated fat. If a triglyceride contains one or more __________ bonds, then it is classified as an unsaturated fat. Lipids are also responsible for a major component of the cell membrane wall that is both attracted to and repelled by water, called __________. The tail of this structure is made of 2 __________, that are water insoluble (hydrophobic). The head of this structure is made of a single __________, that is water soluble (hydrophilic). Proteins building blocks are amino acids that are held together with __________ bonds. These are covalent bonds that link the amino end of one amino acid with the carboxyl end of another. Their overall shape determines their __________. The complex 3D shape of a protein is called a __________. -

Expanding the Genetic Code Lei Wang and Peter G
Reviews P. G. Schultz and L. Wang Protein Science Expanding the Genetic Code Lei Wang and Peter G. Schultz* Keywords: amino acids · genetic code · protein chemistry Angewandte Chemie 34 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim DOI: 10.1002/anie.200460627 Angew. Chem. Int. Ed. 2005, 44,34–66 Angewandte Protein Science Chemie Although chemists can synthesize virtually any small organic molecule, our From the Contents ability to rationally manipulate the structures of proteins is quite limited, despite their involvement in virtually every life process. For most proteins, 1. Introduction 35 modifications are largely restricted to substitutions among the common 20 2. Chemical Approaches 35 amino acids. Herein we describe recent advances that make it possible to add new building blocks to the genetic codes of both prokaryotic and 3. In Vitro Biosynthetic eukaryotic organisms. Over 30 novel amino acids have been genetically Approaches to Protein encoded in response to unique triplet and quadruplet codons including Mutagenesis 39 fluorescent, photoreactive, and redox-active amino acids, glycosylated 4. In Vivo Protein amino acids, and amino acids with keto, azido, acetylenic, and heavy-atom- Mutagenesis 43 containing side chains. By removing the limitations imposed by the existing 20 amino acid code, it should be possible to generate proteins and perhaps 5. An Expanded Code 46 entire organisms with new or enhanced properties. 6. Outlook 61 1. Introduction The genetic codes of all known organisms specify the same functional roles to amino acid residues in proteins. Selectivity 20 amino acid building blocks. These building blocks contain a depends on the number and reactivity (dependent on both limited number of functional groups including carboxylic steric and electronic factors) of a particular amino acid side acids and amides, a thiol and thiol ether, alcohols, basic chain. -

RNA Quality Control in Mirna Expression Analysis
RNA Quality Control in miRNA Expression Analysis Application Note Genomics Authors Abstract Christiane Becker, Michael W. Pfaffl It is generally known that total RNA quality has a distinct influence on the validity and TU München, ZIEL reliability of quantitative PCR results. In addition, the recently published MIQE Physiology Weihenstephan guidelines focus on the pre-PCR steps and state the importance of RNA quality Weihenstephaner Berg 3 assessment. 85384 Freising, Germany Various studies showed the impairing effect of ongoing RNA degradation on mRNA Martina Reiter, Michael W. Pfaffl expression results. Therefore, the verification of RNA integrity prior to downstream BioEPS GmbH, Lise-Meitner-Str. 30 applications like RT-qPCR and mircroarrays is indispensable. A fast and reliable 85354 Freising, Germany assessment of RNA integrity can be done with the Eukaryote Total RNA Nano Assay of the Agilent 2100 Bioanalyzer System. The importance of RNA quality should also be considered in new applications such as the investigation of miRNA expression profiles. With the Agilent Small RNA Assay, Agilent is offering one of the few possibilities for selectively estimating miRNA before expression analysis. However, by now little is known about factors affecting miRNA analysis. Herein, the important impact of total RNA quality on quantification of mRNA and miRNA should be considered. Introduction After extraction, RNAs are very unsta- ble and sensitive to degradation due to 1A the ubiquitous occurrence of nucleases. It is well known that expression profil- ing using microarrays or qPCR is influ- enced by RNA quality1, 2. Therefore, this hazard has to be considered during all preprocessing steps and should be min- imized with cautious handling. -

Molecular Biology and Applied Genetics
MOLECULAR BIOLOGY AND APPLIED GENETICS FOR Medical Laboratory Technology Students Upgraded Lecture Note Series Mohammed Awole Adem Jimma University MOLECULAR BIOLOGY AND APPLIED GENETICS For Medical Laboratory Technician Students Lecture Note Series Mohammed Awole Adem Upgraded - 2006 In collaboration with The Carter Center (EPHTI) and The Federal Democratic Republic of Ethiopia Ministry of Education and Ministry of Health Jimma University PREFACE The problem faced today in the learning and teaching of Applied Genetics and Molecular Biology for laboratory technologists in universities, colleges andhealth institutions primarily from the unavailability of textbooks that focus on the needs of Ethiopian students. This lecture note has been prepared with the primary aim of alleviating the problems encountered in the teaching of Medical Applied Genetics and Molecular Biology course and in minimizing discrepancies prevailing among the different teaching and training health institutions. It can also be used in teaching any introductory course on medical Applied Genetics and Molecular Biology and as a reference material. This lecture note is specifically designed for medical laboratory technologists, and includes only those areas of molecular cell biology and Applied Genetics relevant to degree-level understanding of modern laboratory technology. Since genetics is prerequisite course to molecular biology, the lecture note starts with Genetics i followed by Molecular Biology. It provides students with molecular background to enable them to understand and critically analyze recent advances in laboratory sciences. Finally, it contains a glossary, which summarizes important terminologies used in the text. Each chapter begins by specific learning objectives and at the end of each chapter review questions are also included. -

The Structure and Function of Large Biological Molecules 5
The Structure and Function of Large Biological Molecules 5 Figure 5.1 Why is the structure of a protein important for its function? KEY CONCEPTS The Molecules of Life Given the rich complexity of life on Earth, it might surprise you that the most 5.1 Macromolecules are polymers, built from monomers important large molecules found in all living things—from bacteria to elephants— can be sorted into just four main classes: carbohydrates, lipids, proteins, and nucleic 5.2 Carbohydrates serve as fuel acids. On the molecular scale, members of three of these classes—carbohydrates, and building material proteins, and nucleic acids—are huge and are therefore called macromolecules. 5.3 Lipids are a diverse group of For example, a protein may consist of thousands of atoms that form a molecular hydrophobic molecules colossus with a mass well over 100,000 daltons. Considering the size and complexity 5.4 Proteins include a diversity of of macromolecules, it is noteworthy that biochemists have determined the detailed structures, resulting in a wide structure of so many of them. The image in Figure 5.1 is a molecular model of a range of functions protein called alcohol dehydrogenase, which breaks down alcohol in the body. 5.5 Nucleic acids store, transmit, The architecture of a large biological molecule plays an essential role in its and help express hereditary function. Like water and simple organic molecules, large biological molecules information exhibit unique emergent properties arising from the orderly arrangement of their 5.6 Genomics and proteomics have atoms. In this chapter, we’ll first consider how macromolecules are built. -

Transcription Study Guide This Study Guide Is a Written Version of the Material You Have Seen Presented in the Transcription Unit
Transcription Study Guide This study guide is a written version of the material you have seen presented in the transcription unit. The cell’s DNA contains the instructions for carrying out the work of the cell. These instructions are used by the cell’s protein-making machinery to create proteins. If the cell’s DNA were directly read by the protein-making machinery, however, it could be damaged and the process would be slow and cumbersome. The cell avoids this problem by copying genetic information from its DNA into an intermediate called messenger RNA (mRNA). It is this mRNA that is read by the cell’s protein-making machinery. This process is called transcription. Components In this section you will be introduced to the components involved in the process of RNA synthesis, called transcription. This process requires an enzyme that uses many nucleotide bases to copy the instructions present in DNA into an intermediate messenger RNA molecule. RNA What is RNA? · Like DNA, RNA is a polymer made up of nucleotides. · Unlike DNA, which is composed of two strands of nucleotides twisted together, RNA is single-stranded. It can also sometimes fold into complex three-dimensional structures. · RNA contains the same nucleotides as DNA, with the substitution of uraciluridine (U) for thymidine (T). · RNA is chemically different from DNA so that the cell can easily tell the two apart. · In this animation, you will see one type of RNA, messenger RNA, being put together. · There are three types of RNA: mRNA, which you will read more about; tRNA, which is used in the translation process, and rRNA, which acts as a structural element in the ribosome (a translation component).