Ordinal-Level Phylogenomics of the Arthropod Class
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Fig. Ap. 2.1. Denton Tending His Fairy Shrimp Collection
Fig. Ap. 2.1. Denton tending his fairy shrimp collection. 176 Appendix 1 Hatching and Rearing Back in the bowels of this book we noted that However, salts may leach from soils to ultimately if one takes dry soil samples from a pool basin, make the water salty, a situation which commonly preferably at its deepest point, one can then "just turns off hatching. Tap water is usually unsatis- add water and stir". In a day or two nauplii ap- factory, either because it has high TDS, or because pear if their cysts are present. O.K., so they won't it contains chlorine or chloramine, disinfectants always appear, but you get the idea. which may inhibit hatching or kill emerging If your desire is to hatch and rear fairy nauplii. shrimps the hi-tech way, you should get some As you have read time and again in Chapter 5, guidance from Brendonck et al. (1990) and temperature is an important environmental cue for Maeda-Martinez et al. (1995c). If you merely coaxing larvae from their dormant state. You can want to see what an anostracan is like, buy some guess what temperatures might need to be ap- Artemia cysts at the local aquarium shop and fol- proximated given the sample's origin. Try incu- low directions on the container. Should you wish bation at about 3-5°C if it came from the moun- to find out what's in your favorite pool, or gather tains or high desert. If from California grass- together sufficient animals for a study of behavior lands, 10° is a good level at which to start. -

Phylogenetic Analysis of Anostracans (Branchiopoda: Anostraca) Inferred from Nuclear 18S Ribosomal DNA (18S Rdna) Sequences
MOLECULAR PHYLOGENETICS AND EVOLUTION Molecular Phylogenetics and Evolution 25 (2002) 535–544 www.academicpress.com Phylogenetic analysis of anostracans (Branchiopoda: Anostraca) inferred from nuclear 18S ribosomal DNA (18S rDNA) sequences Peter H.H. Weekers,a,* Gopal Murugan,a,1 Jacques R. Vanfleteren,a Denton Belk,b and Henri J. Dumonta a Department of Biology, Ghent University, Ledeganckstraat 35, B-9000 Ghent, Belgium b Biology Department, Our Lady of the Lake University of San Antonio, San Antonio, TX 78207, USA Received 20 February 2001; received in revised form 18 June 2002 Abstract The nuclear small subunit ribosomal DNA (18S rDNA) of 27 anostracans (Branchiopoda: Anostraca) belonging to 14 genera and eight out of nine traditionally recognized families has been sequenced and used for phylogenetic analysis. The 18S rDNA phylogeny shows that the anostracans are monophyletic. The taxa under examination form two clades of subordinal level and eight clades of family level. Two families the Polyartemiidae and Linderiellidae are suppressed and merged with the Chirocephalidae, of which together they form a subfamily. In contrast, the Parartemiinae are removed from the Branchipodidae, raised to family level (Parartemiidae) and cluster as a sister group to the Artemiidae in a clade defined here as the Artemiina (new suborder). A number of morphological traits support this new suborder. The Branchipodidae are separated into two families, the Branchipodidae and Ta- nymastigidae (new family). The relationship between Dendrocephalus and Thamnocephalus requires further study and needs the addition of Branchinella sequences to decide whether the Thamnocephalidae are monophyletic. Surprisingly, Polyartemiella hazeni and Polyartemia forcipata (‘‘Family’’ Polyartemiidae), with 17 and 19 thoracic segments and pairs of trunk limb as opposed to all other anostracans with only 11 pairs, do not cluster but are separated by Linderiella santarosae (‘‘Family’’ Linderiellidae), which has 11 pairs of trunk limbs. -

The Short-Range Endemic Invertebrate Fauna of the Ravensthorpe Range
THE SHORT-RANGE ENDEMIC INVERTEBRATE FAUNA OF THE RAVENSTHORPE RANGE MARK S. HARVEY MEI CHEN LENG Department of Terrestrial Zoology Western Australian Museum June 2008 2 Executive Summary An intensive survey of short-range endemic invertebrates in the Ravensthorpe Range at 79 sites revealed a small but significant fauna of myriapods and arachnids. Four species of short-range endemic invertebrates were found: • The millipede Antichiropus sp. R • The millipede Atelomastix sp. C • The millipede Atelomastix sp. P • The pseudoscorpion Amblyolpium sp. “WA1” Atelomastix sp. C is the only species found to be endemic to the Ravensthorpe Range and was found at 14 sites. Antichiropus sp. R, Atelomastix sp. P and Amblyolpium sp. “WA1” are also found at nearby locations. Sites of high importance include: site 40 with 7 species; sites 7 and 48 each with 5 species; and sites 18 and 44 each with 4 species. WA Museum - Ravensthorpe Range Survey 3 Introduction Australia contains a multitude of terrestrial invertebrate fauna species, with many yet to be discovered and described. Arthropods alone were recently estimated to consist of approximately more than 250,000 species (Yeates et al. 2004). The majority of these belong to the arthropod classes Insecta and Arachnida, and although many have relatively wide distributions across the landscape, some are highly restricted in range with special ecological requirements. These taxa, termed short-range endemics (Harvey 2002b), are taxa categorised as having poor dispersal abilities and/or requiring very specific habitats, usually with naturally small distributional ranges of less than 10,000 km2 and the following ecological and life-history traits: • poor powers of dispersal; • confinement to discontinuous habitats; • usually highly seasonal, only active during cooler, wetter periods; and • low levels of fecundity. -
Diplopoda, Polydesmida, Paradoxosomatidae) Indicates Multiple Glacial Refugia in Southeastern Australia
A peer-reviewed open-access journal ZooKeys 578:Phylogenetic 15–31 (2016) analysis of the Australian trans-Bass Strait millipede genus Pogonosternum... 15 doi: 10.3897/zookeys.578.8052 RESEARCH ARTICLE http://zookeys.pensoft.net Launched to accelerate biodiversity research Phylogenetic analysis of the Australian trans-Bass Strait millipede genus Pogonosternum (Carl, 1912) (Diplopoda, Polydesmida, Paradoxosomatidae) indicates multiple glacial refugia in southeastern Australia Peter Decker1 1 Senckenberg Museum of Natural History Görlitz, Am Museum 1, 02826 Görlitz, Germany Corresponding author: Peter Decker ([email protected]) Academic editor: R. Mesibov | Received 5 February 2016 | Accepted 10 March 2016 | Published 7 April 2016 http://zoobank.org/B5513E69-0DED-4608-98BD-7ECD401B29E3 Citation: Decker P (2016) Phylogenetic analysis of the Australian trans-Bass Strait millipede genus Pogonosternum (Carl, 1912) (Diplopoda, Polydesmida, Paradoxosomatidae) indicates multiple glacial refugia in southeastern Australia. ZooKeys 578: 15–31. doi: 10.3897/zookeys.578.8052 Abstract This study documents the first detailed phylogenetic analysis of an Australian paradoxosomatid millipede genus. Two mitochondrial genes (partial COI and 16S) as well as partial nuclear 28S rDNA were ampli- fied and sequenced for 41 individuals of the southeastern Australian genus Pogonosternum Jeekel, 1965. The analysis indicates that five species groups ofPogonosternum occur across New South Wales, Victoria and Tasmania: P. nigrovirgatum (Carl, 1912), P. adrianae Jeekel, 1982, P. laetificum Jeekel, 1982 and two undescribed species. P. coniferum (Jeekel, 1965) specimens cluster within P. nigrovirgatum. Most of these five species groups exhibit a pattern of high intraspecific genetic variability and highly local- ized haplotypes, suggesting that they were confined to multiple Pleistocene refugia on the southeastern Australian mainland. -

A New Genus of the Millipede Tribe Brachyiulini (Diplopoda: Julida: Julidae) from the Aegean Region
European Journal of Taxonomy 70: 1-12 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2013.70 www.europeanjournaloftaxonomy.eu 2013 · Lazányi E. & Vagalinski B. This work is licensed under a Creative Commons Attribution 3.0 License. Research article urn:lsid:zoobank.org:pub:5E23F454-2A68-42F6-86FB-7D9952B2FE7B A new genus of the millipede tribe Brachyiulini (Diplopoda: Julida: Julidae) from the Aegean region Eszter LAZÁNYI1,4 & Boyan VAGALINSKI2,3,5 1 Corresponding author: Department of Zoology, Hungarian Natural History Museum, Baross u. 13, H-1088 Budapest, Hungary. E-mail: [email protected] 2 Faculty of Biology, Sofia University, 8 Dragan Tsankov Blvd., 1164 Sofia, Bulgaria. E-mail: [email protected] 3 Institute of Biodiversity and Ecosystem Research, Bulgarian Academy of Sciences, 2 Yurii Gagarin Street, 1113, Sofia, Bulgaria. 4 urn:lsid:zoobank.org:author:02DB48F1-624C-4435-AF85-FA87168CD85A 5 urn:lsid:zoobank.org:author:973B8725-039E-4F29-8D73-96A7F52CF934 Abstract. A new genus of the julid tribe Brachyiulini, Enghophyllum gen. nov., is described, comprising two species from Greece. The type-species, E. naxium (Verhoeff, 1901) comb. nov. (ex Megaphyllum Verhoeff, 1894), appears to be rather widespread in the Aegean: it is known from Antiparos Island and Naxos Island (the type locality), both in the Cyclades, as well as East Mavri Islet, Dodecanese Archipelago (new record). The vulva of E. naxium is described for the first time. In addition,E. sifnium gen. et sp. nov. is described based on a single adult male from Sifnos Island, Cyclades. The new genus is distinct from other genera of the Brachyiulini mainly by its peculiar gonopod structure, apparently disjunct and at least mostly apomorphous: (1) promeres broad, shield-like, in situ protruding mostly posteriad, completely covering the opisthomeres and gonopodal sinus; (2) transverse muscles and coxal apodemes of promere fully reduced; (3) opisthomere with three differentiated processes, i.e., lateral, basal posterior and apical posterior; (4) solenomere rather simple, tubular. -

Type and Figured Fossils in the Worthen Collection at the Illinois
s Cq&JI ^XXKUJtJLI 14oGS: CIR 524 c, 2 TYPE AND FIGURED FOSSILS IN THE WORTHEN COLLECTION AT THE ILLINOIS STATE GEOLOGICAL SURVEY Lois S. Kent GEOLOGICAL ILLINOIS Illinois Department of Energy and Natural Resources, STATE GEOLOGICAL SURVEY DIVISION CIRCULAR 524 1982 COVER: This portrait of Amos Henry Worthen is from a print presented to me by Worthen's great-grandson, Arthur C. Brookley, Jr., at the time he visited the Illinois State Geological Survey in the late 1950s or early 1960s. The picture is the same as that published in connection with the memorial to Worthen in the appendix to Vol. 8 of the Geological Survey of Illinois, 1890. -LSK Kent, Lois S., Type and figured fossils in the Worthen Collection at the Illinois State Geological Survey. — Champaign, III. : Illinois State Geological Survey, 1982. - 65 p. ; 28 cm. (Circular / Illinois State Geological Survey ; 524) 1. Paleontology. 2. Catalogs and collections. 3. Worthen Collection. I. Title. II. Series. Editor: Mary Clockner Cover: Sandra Stecyk Printed by the authority of the State of Illinois/1982/2500 II I IHOI'.MAII '.I 'II Of.ir.AI MIHVI y '> 300 1 00003 5216 TYPE AND FIGURED FOSSILS IN THE WORTHEN COLLECTION AT THE ILLINOIS STATE GEOLOGICAL SURVEY Lois S. Kent | CIRCULAR 524 1982 ILLINOIS STATE GEOLOGICAL SURVEY Robert E. Bergstrom, Acting Chief Natural Resources Building, 615 East Peabody Drive, Champaign, IL 61820 TYPE AND FIGURED FOSSILS IN THE WORTHEN COLLECTION AT THE ILLINOIS STATE GEOLOGICAL SURVEY CONTENTS Acknowledgments 2 Introduction 2 Organization of the catalog 7 Notes 8 References 8 Fossil catalog 13 ABSTRACT This catalog lists all type and figured specimens of fossils in the part of the "Worthen Collection" now housed at the Illinois State Geological Survey in Champaign, Illinois. -

MYRIAPODS 767 Volume 2 (M-Z), Pp
In: R. Singer, (ed.), 1999. Encyclopedia of Paleontology, MYRIAPODS 767 volume 2 (M-Z), pp. 767-775. Fitzroy Dearborn, London. MYRIAPODS JVlyriapods are many-legged, terrestrial arthropods whose bodies groups, the Trilobita, Chelicerata, Crustacea, and the Uniramia, the are divided into two major parts, a head and a trunk. The head last consisting of the Myriapoda, Hexapoda, and Onychophora (vel- bears a single pair of antennae, highly differentiated mandibles (or vet worms). However, subsequent structural and molecular evidence jaws), and at least one pair of maxillary mouthparts; the trunk indicates that there are several characters uniting major arthropod region consists of similar "metameres," each of which is a func- taxa. Moreover, paleobiologic, embryologie, and other evidence tional segment that bears one or two pairs of appendages. Gas demonstrates that myriapods and hexapods are fiindamentally exchange is accomplished by tracheae•a branching network of polyramous, having two major articulating appendages per embry- specialized tubules•although small forms respire through the ological body segment, like other arthropods. body wall. Malpighian organs are used for excretion, and eyes con- A fourth proposal (Figure ID) suggests that myriapods are sist of clusters of simple, unintegrated, light-sensitive elements an ancient, basal arthropod lineage, and that the Hexapoda that are termed ommatidia. These major features collectively char- emerged as an independent, relatively recent clade from a rather acterize the five major myriapod clades: Diplopoda (millipeds), terminal crustacean lineage, perhaps the Malacostraca, which con- Chilopoda (centipeds), Pauropoda (pauropods), Symphyla (sym- tains lobsters and crabs (Ballard et al. 1992). Because few crusta- phylans), and Arthropleurida (arthropleurids). Other features cean taxa were examined in this analysis, and due to the Cambrian indicate differences among these clades. -

Mandrillus Leucophaeus Poensis)
Ecology and Behavior of the Bioko Island Drill (Mandrillus leucophaeus poensis) A Thesis Submitted to the Faculty of Drexel University by Jacob Robert Owens in partial fulfillment of the requirements for the degree of Doctor of Philosophy December 2013 i © Copyright 2013 Jacob Robert Owens. All Rights Reserved ii Dedications To my wife, Jen. iii Acknowledgments The research presented herein was made possible by the financial support provided by Primate Conservation Inc., ExxonMobil Foundation, Mobil Equatorial Guinea, Inc., Margo Marsh Biodiversity Fund, and the Los Angeles Zoo. I would also like to express my gratitude to Dr. Teck-Kah Lim and the Drexel University Office of Graduate Studies for the Dissertation Fellowship and the invaluable time it provided me during the writing process. I thank the Government of Equatorial Guinea, the Ministry of Fisheries and the Environment, Ministry of Information, Press, and Radio, and the Ministry of Culture and Tourism for the opportunity to work and live in one of the most beautiful and unique places in the world. I am grateful to the faculty and staff of the National University of Equatorial Guinea who helped me navigate the geographic and bureaucratic landscape of Bioko Island. I would especially like to thank Jose Manuel Esara Echube, Claudio Posa Bohome, Maximilliano Fero Meñe, Eusebio Ondo Nguema, and Mariano Obama Bibang. The journey to my Ph.D. has been considerably more taxing than I expected, and I would not have been able to complete it without the assistance of an expansive list of people. I would like to thank all of you who have helped me through this process, many of whom I lack the space to do so specifically here. -

Motion Control of Daphnia Magna by Blue LED Light
Motion Control of Daphnia magna by Blue LED Light Akitoshi Itoha,* and Hirotomo Hisamab aDepartment of Mechanical Engineering, Tokyo Denki University, Tokyo, Japan bGraduate School Student, Dept. of Mech. Eng., Tokyo Denki University, Tokyo, Japan Abstract—Daphnia magna show strong positive protests, and similar studies have not yet been phototaxis to blue light. Here, we investigate the conducted on multicelluar motile plankton species. effectiveness of behavior control of D. magna by blue Multicellular motile planktons have larger bodies than light irradiation for their use as bio-micromachines. D. protists and are more highly evolved, enhancing their magna immediately respond by swimming toward blue ability to carry out more complex tasks provided that LED light sources. The behavior of individual D. they can be controlled. Tools may be easily attached to magna was controlled by switching on the LED placed their exoskeletons, and the small (body length < 0.5 at 15° intervals around a shallow Petri-dish to give a mm), motile zooplankton or their juvenile instars may target direction. The phototaxic controllability of have potential as bio-micromachines. Daphnia was much better than the galvanotactic controllability of Paramecium. II. TAXES OF MULTICELLULAR MOTILE ZOOPLANKTON Index Terms— Daphnia, Bio-micromachine, Generally, rearing zooplankton is more difficult than Phototaxis, Motion Control motile protists due to the requirements for also growing natural food (mainly phytoplankton) in culture and to I. INTRODUCTION the lack of suitable artificial diets. First, we collected Studies to use microorganisms as bio-micromachines five species, comprising 4 Branchiopoda (Daphnia was first investigated in 1986 as presented by Fearing magna, Moina sp., Bosmina sp., Scapholeberis sp.), [1]. -
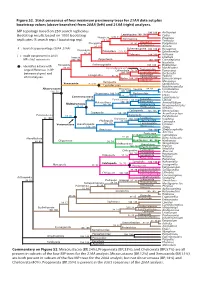
Figure S2. Strict Consensus of Four Maximum Parsimony Trees for 21AA Data Set Plus Bootstrap Values (Above Branches) from 20AA (Left) and 21AA (Right) Analyses
Figure S2. Strict consensus of four maximum parsimony trees for 21AA data set plus bootstrap values (above branches) from 20AA (left) and 21AA (right) analyses. MP topology based on 250 search replicates. 100 100 Antheraea Bootstrap results based on 1000 bootstrap Lepidoptera 100 100 Cydia Neoptera [23] 44 Dermaptera Prodoxus replicates (5 search reps / bootstrap rep). 68 66 Forficula Blattodea Pterygota 92 96 Periplaneta Orthoptera 96 97 Acheta # : bootstrap percentage (20AA 21AA) Ephemeroptera Dicondylia 100 100 Hexagenia Paleoptera [37] 52 Ephemerella 98 98 Odonata 100 100 Ischnura [ ] : node not present in 20AA Insecta Libellula MP strict consensus 100 100 Zygentoma 100 100 Ctenolepisma Nicoletia Archaeognatha : Identifies 6 taxa with Hexapoda 100 100 Pedetontus 89 86 Machiloides Entomobryomorpha Tomoceridae large differences in BP Collembola 70 76 Tomocerus between degen1 and 100 100 Entomobryidae Orchesella Entognatha 82 82 Poduridae Podura 20AA analyses. Diplura 100 100 Campodeidae Japygidae Eumesocampa Remipedia Metajapyx Xenocarida [31] 51 Speleonectes Cephalocarida Hutchinsoniella Altocrustacea Thoracica Sessilia 88 83 Semibalanus [14] 31 100 100 Chthamalus Thecostraca 100 100 Pedunculata Lepas Communostraca Rhizocephala Loxothylacus 96 89 Eumalacostraca 84 85 Eucarida Libinia Malacostraca 100 100 Peracarida Armadillidium Multicrustacea 100 100 Hoplocarida 69 84 Neogonodactylus Phyllocarida Nebalia Cyclopoida 100 100 Mesocyclops Copepoda 100 100 Acanthocyclops Pancrustacea 65 82 Calanoida Eurytemora 96 100 Diplostraca 100 100 -

Exploring Phylogenomic Relationships Within Myriapoda: Should High Matrix Occupancy Be the Goal?
bioRxiv preprint doi: https://doi.org/10.1101/030973; this version posted November 9, 2015. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Exploring phylogenomic relationships within Myriapoda: should high matrix occupancy be the goal? ROSA FERNÁNDEZ1, GREGORY D. EDGECOMBE2 AND GONZALO GIRIBET1 1Museum of Comparative Zoology & Department of Organismic and Evolutionary Biology, Harvard University, 26 Oxford Street, Cambridge, MA 02138, USA 2Department of Earth Sciences, The Natural History Museum, Cromwell Road, London SW7 5BD, UK 1 bioRxiv preprint doi: https://doi.org/10.1101/030973; this version posted November 9, 2015. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract.—Myriapods are one of the dominant terrestrial arthropod groups including the diverse and familiar centipedes and millipedes. Although molecular evidence has shown that Myriapoda is monophyletic, its internal phylogeny remains contentious and understudied, especially when compared to those of Chelicerata and Hexapoda. Until now, efforts have focused on taxon sampling (e.g., by including a handful of genes in many species) or on maximizing matrix occupancy (e.g., by including hundreds or thousands of genes in just a few species), but a phylogeny maximizing sampling at both levels remains elusive. In this study, we analyzed forty Illumina transcriptomes representing three myriapod classes (Diplopoda, Chilopoda and Symphyla); twenty-five transcriptomes were newly sequenced to maximize representation at the ordinal level in Diplopoda and at the family level in Chilopoda. -

'Marri Millipede' Antichiropus Variabilis
RECORDS OF THE WESTERN AUSTRALIAN MUSEUM 26 087–093 (2010) Optimised captive husbandry conditions for the Western Australian ‘Marri Millipede’ Antichiropus variabilis (Diplopoda: Polydesmida: Paradoxosomatidae), with notes on natural history and tissue preservation techniques Janine M. Wojcieszek1, Mark S. Harvey2,1 and Michael G. Rix2 1Centre for Evolutionary Biology, School of Animal Biology M092, University of Western Australia, 35 Stirling Highway, Crawley, Perth, Western Australia 6009, Australia. Email: [email protected] 2Department of Terrestrial Zoology, Western Australian Museum, Locked Bag 49, Welshpool D.C., Perth, Western Australia 6986, Australia. ABSTRACT – The millipede genus Antichiropus Attems, 1911, is extremely diverse and the majority of species are endemic to south-western Western Australia. Very little is known about the general biology of species of Antichiropus; however, these millipedes are becoming useful models for studies of speciation and sexual selection, and remain central to SRE-based conservation planning for government and industry in the expanding resources sector of Western Australia. This paper details optimised captive husbandry conditions and observations made regarding the natural history of one species – Antichiropus variabilis – following three years of fi eld collecting and laboratory-based behavioural and molecular research. INTRODUCTION found in Marri (Corymbia calophylla) and Jarrah (Eucalyptus marginata) forests along much of the The genus Antichiropus is an extremely diverse Darling Escarpment east of Perth (Harvey 2002) (Figure group of largely short-range endemic (SRE) millipedes 2). Recent research investigating sexual selection (Harvey 2002), occurring in south-western Western in A. variabilis required the development of captive Australia and western South Australia (Harvey and husbandry procedures for laboratory experiments, and Mesibov 2007).