Protozoologica
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

A Revised Classification of Naked Lobose Amoebae (Amoebozoa
Protist, Vol. 162, 545–570, October 2011 http://www.elsevier.de/protis Published online date 28 July 2011 PROTIST NEWS A Revised Classification of Naked Lobose Amoebae (Amoebozoa: Lobosa) Introduction together constitute the amoebozoan subphy- lum Lobosa, which never have cilia or flagella, Molecular evidence and an associated reevaluation whereas Variosea (as here revised) together with of morphology have recently considerably revised Mycetozoa and Archamoebea are now grouped our views on relationships among the higher-level as the subphylum Conosa, whose constituent groups of amoebae. First of all, establishing the lineages either have cilia or flagella or have lost phylum Amoebozoa grouped all lobose amoe- them secondarily (Cavalier-Smith 1998, 2009). boid protists, whether naked or testate, aerobic Figure 1 is a schematic tree showing amoebozoan or anaerobic, with the Mycetozoa and Archamoe- relationships deduced from both morphology and bea (Cavalier-Smith 1998), and separated them DNA sequences. from both the heterolobosean amoebae (Page and The first attempt to construct a congruent molec- Blanton 1985), now belonging in the phylum Per- ular and morphological system of Amoebozoa by colozoa - Cavalier-Smith and Nikolaev (2008), and Cavalier-Smith et al. (2004) was limited by the the filose amoebae that belong in other phyla lack of molecular data for many amoeboid taxa, (notably Cercozoa: Bass et al. 2009a; Howe et al. which were therefore classified solely on morpho- 2011). logical evidence. Smirnov et al. (2005) suggested The phylum Amoebozoa consists of naked and another system for naked lobose amoebae only; testate lobose amoebae (e.g. Amoeba, Vannella, this left taxa with no molecular data incertae sedis, Hartmannella, Acanthamoeba, Arcella, Difflugia), which limited its utility. -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

Diagnosis of Infections Caused by Pathogenic Free-Living Amoebae
Virginia Commonwealth University VCU Scholars Compass Microbiology and Immunology Publications Dept. of Microbiology and Immunology 2009 Diagnosis of Infections Caused by Pathogenic Free- Living Amoebae Bruno da Rocha-Azevedo Virginia Commonwealth University Herbert B. Tanowitz Albert Einstein College of Medicine Francine Marciano-Cabral Virginia Commonwealth University Follow this and additional works at: http://scholarscompass.vcu.edu/micr_pubs Part of the Medicine and Health Sciences Commons Copyright © 2009 Bruno da Rocha-Azevedo et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Downloaded from http://scholarscompass.vcu.edu/micr_pubs/9 This Article is brought to you for free and open access by the Dept. of Microbiology and Immunology at VCU Scholars Compass. It has been accepted for inclusion in Microbiology and Immunology Publications by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Hindawi Publishing Corporation Interdisciplinary Perspectives on Infectious Diseases Volume 2009, Article ID 251406, 14 pages doi:10.1155/2009/251406 Review Article Diagnosis of Infections Caused by Pathogenic Free-Living Amoebae Bruno da Rocha-Azevedo,1 Herbert B. Tanowitz,2 and Francine Marciano-Cabral1 1 Department of Microbiology and Immunology, Virginia Commonwealth University School of Medicine, Richmond, VA 23298, USA 2 Department of Pathology, Albert Einstein College of Medicine, Bronx, NY 10461, USA Correspondence should be addressed to Francine Marciano-Cabral, [email protected] Received 25 March 2009; Accepted 5 June 2009 Recommended by Louis M. Weiss Naegleria fowleri, Acanthamoeba spp., Balamuthia mandrillaris,andSappinia sp. -

Author's Manuscript (764.7Kb)
1 BROADLY SAMPLED TREE OF EUKARYOTIC LIFE Broadly Sampled Multigene Analyses Yield a Well-resolved Eukaryotic Tree of Life Laura Wegener Parfrey1†, Jessica Grant2†, Yonas I. Tekle2,6, Erica Lasek-Nesselquist3,4, Hilary G. Morrison3, Mitchell L. Sogin3, David J. Patterson5, Laura A. Katz1,2,* 1Program in Organismic and Evolutionary Biology, University of Massachusetts, 611 North Pleasant Street, Amherst, Massachusetts 01003, USA 2Department of Biological Sciences, Smith College, 44 College Lane, Northampton, Massachusetts 01063, USA 3Bay Paul Center for Comparative Molecular Biology and Evolution, Marine Biological Laboratory, 7 MBL Street, Woods Hole, Massachusetts 02543, USA 4Department of Ecology and Evolutionary Biology, Brown University, 80 Waterman Street, Providence, Rhode Island 02912, USA 5Biodiversity Informatics Group, Marine Biological Laboratory, 7 MBL Street, Woods Hole, Massachusetts 02543, USA 6Current address: Department of Epidemiology and Public Health, Yale University School of Medicine, New Haven, Connecticut 06520, USA †These authors contributed equally *Corresponding author: L.A.K - [email protected] Phone: 413-585-3825, Fax: 413-585-3786 Keywords: Microbial eukaryotes, supergroups, taxon sampling, Rhizaria, systematic error, Excavata 2 An accurate reconstruction of the eukaryotic tree of life is essential to identify the innovations underlying the diversity of microbial and macroscopic (e.g. plants and animals) eukaryotes. Previous work has divided eukaryotic diversity into a small number of high-level ‘supergroups’, many of which receive strong support in phylogenomic analyses. However, the abundance of data in phylogenomic analyses can lead to highly supported but incorrect relationships due to systematic phylogenetic error. Further, the paucity of major eukaryotic lineages (19 or fewer) included in these genomic studies may exaggerate systematic error and reduces power to evaluate hypotheses. -
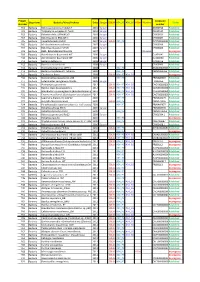
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

EVALUATION of the P45 MOBILE INTEGRATIVE ELEMENT and ITS ROLE IN
EVALUATION OF THE p45 MOBILE INTEGRATIVE ELEMENT AND ITS ROLE IN Legionella pneumophila VIRULENCE A Dissertation by LANETTE M. CHRISTENSEN Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Jeffrey D. Cirillo Committee Members, James Samuel Jon Skare Farida Sohrabji Head of Program, Warren Zimmer May 2018 Major Subject: Medical Sciences Copyright 2018 Lanette Christensen ABSTRACT Legionella pneumophila are aqueous environmental bacilli that live within protozoal species and cause a potentially fatal form of pneumonia called Legionnaires’ disease. Not all L. pneumophila strains have the same capacity to cause disease in humans. The majority of strains that cause clinically relevant Legionnaires’ disease harbor the p45 mobile integrative genomic element. Contribution of the p45 element to L. pneumophila virulence and ability to withstand environmental stress were addressed in this study. The L. pneumophila Philadelphia-1 (Phil-1) mobile integrative element, p45, was transferred into the attenuated strain Lp01 via conjugation, designating p45 an integrative conjugative element (ICE). The resulting trans-conjugate, Lp01+p45, was compared with strains Phil-1 and Lp01 to assess p45 in virulence using a guinea pig model infected via aerosol. The p45 element partially recovered the loss of virulence in Lp01 compared to that of Phil-1 evident in morbidity, mortality, and bacterial burden in the lungs at the time of death. This phenotype was accompanied by enhanced expression of type II interferon in the lungs and spleens 48 hours after infection, independent of bacterial burden. -

Protistology Light-Microscopic Morphology and Ultrastructure Of
Protistology 13 (1), 26–35 (2019) Protistology Light-microscopic morphology and ultrastructure of Polychaos annulatum (Penard, 1902) Smirnov et Goodkov, 1998 (Amoebozoa, Tubulinea, Euamo- ebida), re-isolated from the surroundings of St. Petersburg (Russia) Oksana Kamyshatskaya1,2, Yelisei Mesentsev1, Ludmila Chistyakova2 and Alexey Smirnov1 1 Department of Invertebrate Zoology, Faculty of Biology, St. Petersburg State University, Universitetskaya nab. 7/9, 199034 St. Petersburg, Russia 2 Core Facility Center “Culturing of microorganisms”, Research park of St. Petersburg State Univeristy, St. Petersburg State University, Botanicheskaya St., 17A, 198504, Peterhof, St. Petersburg, Russia | Submitted December 15, 2018 | Accepted January 21, 2019 | Summary We isolated the species Polychaos annulatum (Penard, 1902) Smirnov et Goodkov, 1998 from a freshwater habitat in the surrounding of Saint-Petersburg. The previous re-isolation of this species took place in 1998; at that time the studies of its light- microscopic morphology were limited with the phase contrast optics, and the electron- microscopic data were obtained using the traditional glutaraldehyde fixation, preceded with prefixation and followed by postfixation with osmium tetroxide. In the present paper, we provide modern DIC images of P. annulatum. Using the fixation protocol that includes a mixture of the glutaraldehyde and paraformaldehyde we were able to obtain better fixation quality for this species. We provide some novel details of its locomotive morphology, nuclear morphology, and ultrastructure. The present finding evidence that P. annulatum is a widely distributed species that could be isolated from a variety of freshwater habitats. Key words: amoebae, Polychaos, ultrastructure, Amoebozoa Introduction Amoeba proteus). These amoebae usually have a relatively large size, exceeding hundred of microns, The largest species of naked lobose amoebae they produce broad, thick pseudopodia with (gymnamoebae) – members of the family Amoe- smooth outlines (lobopodia). -

Bacterial Species Exclusively Identified in Male Subjects
Supplementary Table 1: Bacterial species exclusively identified in male subjects Acholeplasma ales Clostridium magnum Lactobacillus ruminis Pseudomonas collierea Acholeplasma cavigenitalium Clostridium malenominatum Lactobacillus salivarius Pseudomonas jinjuensis Acholeplasma equifetale Clostridium nitrophenolicum Lactobacillus versmoldensis Pseudomonas luteola Acholeplasma granularum Clostridium paraputrificum Lactococcus lactis Pseudomonas panipatensis Acholeplasma hippikon Clostridium perfringens Legionella fallonii Pseudomonas poae Acidaminobacter hydrogenoformans Clostridium proteolyticum Legionella taurinensis Pseudomonas tropicalis Acidaminococcus intestini Clostridium proteolyticus Legionella worsleiensis Pseudonocardia acaciae Clostridium Acidiphilium acidophilum saccharoperbutylacetonicum Lentzea californiensis Pseudonocardia asaccharolytica Pseudonocardia Acidiphilium organovorum Clostridium sulfidigenes Leptotrichia buccalis hydrocarbonoxydans Acidiphilium symbioticum Clostridium termitidis Leptotrichia shahii Pseudoxanthomonas mexicana Acidisoma tundrae Clostridium tetani Leptotrichia wadei Pullulanibacillus naganoensis Acidovorax facilis Clostridium thermoalcaliphilum Leucobacter albus Rhizobium mesoamericanum Acinetobacter indicus Clostridium tunisiense Lewinella marina Rhodanobacter thiooxydans Acinetobacter venetianus Cohnella fontinalis Luteimonas aquatica Rhodobacter blasticus Actinoallomurus luridus Cohnella laeviribosi Luteimonas mephitis Rhodobacter gluconicum Actinobacillus pleuropneumoniae Collinsella stercoris -

Three Unusual New Legionella Species
International Journal of Systematic and Evolutionary Microbiology (2001), 51, 1151–1160 Printed in Great Britain Legionella drozanskii sp. nov., Legionella rowbothamii sp. nov. and Legionella fallonii sp. nov.: three unusual new Legionella species 1 Division of Life Sciences, Adenike A. Adeleke,1 Barry S. Fields,2 Robert F. Benson,2 King’s College London, 2 2 3 Franklin-Wilkins Building, Maryam I. Daneshvar, Janet M. Pruckler, Rodney M. Ratcliff, 150 Stamford Street, Timothy G. Harrison,4 Robbin S. Weyant,2 Richard J. Birtles,5 London SE1 8WA, UK Didier Raoult6 and Mahmoud A. Halablab1 2 Division of Bacterial and Mycotic Diseases, Centers for Disease Control and Author for correspondence: Mahmoud A. Halablab. Tel: j44 207 848 4281. Fax: j44 207 848 4500. Prevention (CDC), Atlanta, e-mail: mahmoud.halablab!kcl.ac.uk GA 30333, USA 3 Infectious Diseases Laboratories, Institute of Seven strains of Legionella-like amoebal pathogens (LLAPs) were characterized Medical and Veterinary on the basis of their cultural and staining characteristics, biochemical Science, Frome Road, reactions, serology, cellular fatty acids (CFAs), isoprenoid quinone Adelaide, SA, 5000, Australia composition, total DNA relatedness, analysis of 16S rRNA and macrophage infectivity potentiator (mip) gene sequence analyses. All seven strains 4 PHLS Central Public Health Laboratory, Colindale, exhibited limited growth on buffered charcoal yeast extract α (BCYE) agar, London NW9 5HT, UK required cysteine for growth and contained branched-chain CFAs and quinones 5 Department of typical of Legionella species. The bacilli were Gram-negative and catalase- Microbiology, University of positive. There were varying degrees of serological cross-reactions between Bristol, Bristol BS8 1TD, UK these LLAP strains and other previously described Legionella species. -

Protistology Re Description of Vannella Mira Schaeffer 1926
Protistology 2 (3), 178–184 (2002) Protistology Redescription of Vannella mira Schaeffer 1926 (Gymnamoebia, Vannellidae), an often mentioned but poorly known amoebae species Alexey V. Smirnov Marine Biological Laboratory University of Copenhagen, Helsingor, Denmark and Department of Invertebrate Zoology, Faculty of Biology and Soil Sci., St.Petersburg State University Summary Amoebae species Vannella mira Schaeffer 1926 was relatively well described and has often been discovered in various regional faunas. However, there is still no clear understanding as to what we really mean by the name “Vannella mira”. A.A. Schaeffer described this species from marine environment in 1926 (as Flabellula mira), but further researchers applied this name for several brackish and freshwater isolates. F.C. Page redescribed “V. mira” in 1968 from freshwater and in 1976 listed it among freshwater amoebae. Perhaps, it was a misidentification. In 1988 he revised his opinion stating that “long experience has failed to reveal any other gymnamoeba occurring in both fresh and salt water” and applied a specific name Vannella cirifera Frenzel 1892 to the isolate that he previously called V. mira. However, various authors used the name V. mira for different marine and freshwater isolates even after the renaming. The present paper describes a marine amoeba isolated from cyanobacterial mats in France (Camarque) that corresponds in full to Schaeffer’s initial description of “Flabellula mira”. New type material is established for the species V. mira Schaeffer 1926; it remains a type species of the genus Vannella. Key words: amoebae, Vannella, Lobosea, Gymnamoebia, morphology, systematics, Vannellidae Introduction shape always present in endoplasm. No vacuoles. -

Comparative Analyses of Legionella Species Identifies Genetic Features
Gomez-Valero et al. Genome Biology 2014, 15:505 http://genomebiology.com/2014/15/11/505 RESEARCH Open Access Comparative analyses of Legionella species identifies genetic features of strains causing Legionnaires’ disease Laura Gomez-Valero1,2*, Christophe Rusniok1,2, Monica Rolando1,2, Mario Neou1,2, Delphine Dervins-Ravault1,2, Jasmin Demirtas1,2, Zoe Rouy9, Robert J Moore10, Honglei Chen10, Nicola K Petty3, Sophie Jarraud4,5, Jerome Etienne4,5, Michael Steinert6, Klaus Heuner7, Simonetta Gribaldo8, Claudine Médigue9, Gernot Glöckner11, Elizabeth L Hartland12 and Carmen Buchrieser1* Abstract Background: The genus Legionella comprises over 60 species. However, L. pneumophila and L. longbeachae alone cause over 95% of Legionnaires’ disease. To identify the genetic bases underlying the different capacities to cause disease we sequenced and compared the genomes of L. micdadei, L. hackeliae and L. fallonii (LLAP10), which are all rarely isolated from humans. Results: We show that these Legionella species possess different virulence capacities in amoeba and macrophages, correlating with their occurrence in humans. Our comparative analysis of 11 Legionella genomes belonging to five species reveals highly heterogeneous genome content with over 60% representing species-specific genes; these comprise a complete prophage in L. micdadei, the first ever identified in a Legionella genome. Mobile elements are abundant in Legionella genomes; many encode type IV secretion systems for conjugative transfer, pointing to their importance for adaptation of the genus. The Dot/Icm secretion system is conserved, although the core set of substrates is small, as only 24 out of over 300 described Dot/Icm effector genes are present in all Legionella species. We also identified new eukaryotic motifs including thaumatin, synaptobrevin or clathrin/coatomer adaptine like domains.