To Measure ADAM10 and ADAM17 Sheddase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ACE2/ADAM17/TMPRSS2 Interplay May Be the Main Risk Factor for COVID-19
REVIEW published: 07 October 2020 doi: 10.3389/fimmu.2020.576745 ACE2/ADAM17/TMPRSS2 Interplay May Be the Main Risk Factor for COVID-19 † † Donato Zipeto 1 , Julys da Fonseca Palmeira 2 , Gustavo A. Argañaraz 2 and Enrique R. Argañaraz 2* 1 Department of Neuroscience, Biomedicine and Movement Sciences, University of Verona, Verona, Italy, 2 Laboratory of Molecular Neurovirology, Faculty of Health Science, University of Bras´ılia, Brasilia, Brazil The Coronavirus Disease 2019 (COVID-19) has already caused hundreds of thousands of deaths worldwide in a few months. Cardiovascular disease, hypertension, diabetes and chronic lung disease have been identified as the main COVID-19 comorbidities. Moreover, Edited by: despite similar infection rates between men and women, the most severe course of the Yolande Richard, Institut National de la Sante´ et de la disease is higher in elderly and co-morbid male patients. Therefore, the occurrence of Recherche Me´ dicale (INSERM), specific comorbidities associated with renin–angiotensin system (RAS) imbalance France mediated by the interaction between angiotensin-converting enzyme 2 (ACE2) and Reviewed by: desintegrin and metalloproteinase domain 17 (ADAM17), along with specific genetic Andreas Ludwig, RWTH Aachen University, factors mainly associated with type II transmembrane serine protease (TMPRSS2) Germany expression, could be decisive for the clinical outcome of COVID-19. Indeed, the Elena Ciaglia, — University of Salerno, Italy exacerbated ADAM17 mediated ACE2, TNF-a, and IL-6R secretion emerges as a Rumi Ueha, possible underlying mechanism for the acute inflammatory immune response and the University of Tokyo, Japan activation of the coagulation cascade. Therefore, in this review, we focus on the main *Correspondence: pathophysiological aspects of ACE2, ADAM17, and TMPRSS2 host proteins in COVID- Enrique R. -

Neprilysin Is Required for Angiotensin-(1-7)
Page 1 of 39 Diabetes NEPRILYSIN IS REQUIRED FOR ANGIOTENSIN-(1-7)’S ABILITY TO ENHANCE INSULIN SECRETION VIA ITS PROTEOLYTIC ACTIVITY TO GENERATE ANGIOTENSIN-(1-2) Gurkirat S. Brara, Breanne M. Barrowa, Matthew Watsonb, Ryan Griesbachc, Edwina Chounga, Andrew Welchc, Bela Ruzsicskad, Daniel P. Raleighb, Sakeneh Zraikaa,c aVeterans Affairs Puget Sound Health Care System, Seattle, WA 98108, United States bDepartment of Chemistry, Stony Brook University, Stony Brook, NY 11794, United States cDivision of Metabolism, Endocrinology and Nutrition, Department of Medicine, University of Washington, Seattle, WA 98195, United States dInstitute for Chemical Biology and Drug Discovery, Stony Brook University, Stony Brook, NY 11794, United States Short Title: Angiotensin-(1-7) and insulin secretion Word count: 3997; Figure count: 8 main (plus 3 Online Suppl.); Table count: 1 Online Suppl. Correspondence to: Sakeneh Zraika, PhD 1660 South Columbian Way (151) Seattle, WA, United States Tel: 206-768-5391 / Fax: 206-764-2164 Email: [email protected] 1 Diabetes Publish Ahead of Print, published online May 30, 2017 Diabetes Page 2 of 39 ABSTRACT Recent work has renewed interest in therapies targeting the renin-angiotensin system (RAS) to improve β-cell function in type 2 diabetes. Studies show that generation of angiotensin-(1-7) by angiotensin converting enzyme 2 (ACE2) and its binding to the Mas receptor (MasR) improves glucose homeostasis, partly by enhancing glucose-stimulated insulin secretion (GSIS). Thus, islet ACE2 upregulation is viewed as a desirable therapeutic goal. Here, we show that although endogenous islet ACE2 expression is sparse, its inhibition abrogates angiotensin-(1-7)-mediated GSIS. However, a more widely expressed islet peptidase, neprilysin, degrades angiotensin-(1-7) into several peptides. -

Advanced Glycation End Products Evoke Endothelial Cell Damage By
Ishibashi et al. Cardiovascular Diabetology 2013, 12:125 CARDIO http://www.cardiab.com/content/12/1/125 VASCULAR DIABETOLOGY ORIGINAL INVESTIGATION Open Access Advanced glycation end products evoke endothelial cell damage by stimulating soluble dipeptidyl peptidase-4 production and its interaction with mannose 6-phosphate/insulin- like growth factor II receptor Yuji Ishibashi1, Takanori Matsui1, Sayaka Maeda1, Yuichiro Higashimoto2 and Sho-ichi Yamagishi1* Abstract Background: Advanced glycation end products (AGEs) and receptor RAGE interaction play a role in diabetic vascular complications. Inhibition of dipeptidyl peptidase-4 (DPP-4) is a potential therapeutic target for type 2 diabetes. However, the role of DPP-4 in AGE-induced endothelial cell (EC) damage remains unclear. Methods: In this study, we investigated the effects of DPP-4 on reactive oxygen species (ROS) generation and RAGE gene expression in ECs. We further examined whether an inhibitor of DPP-4, linagliptin inhibited AGE-induced soluble DPP-4 production, ROS generation, RAGE, intercellular adhesion molecule-1 (ICAM-1) and plasminogen activator inhibitor-1 (PAI-1) gene expression in ECs. Results: DPP-4 dose-dependently increased ROS generation and RAGE gene expression in ECs, which were prevented by linagliptin. Mannose 6-phosphate (M6P) and antibodies (Ab) raised against M6P/insulin-like growth factor II receptor (M6P/IGF-IIR) completely blocked the ROS generation in DPP-4-exposed ECs, whereas surface plasmon resonance revealed that DPP-4 bound to M6P/IGF-IIR at the dissociation constant of 3.59 x 10-5 M. AGEs or hydrogen peroxide increased soluble DPP-4 production by ECs, which was prevented by N-acetylcysteine, RAGE-Ab or linagliptin. -

Intestinal Tumorigenesis Is Suppressed in Mice Lacking the Metalloproteinase Matrilysin (Knockout͞min͞colon Cancer͞extracellular Matrix͞apc)
Proc. Natl. Acad. Sci. USA Vol. 94, pp. 1402–1407, February 1997 Medical Sciences Intestinal tumorigenesis is suppressed in mice lacking the metalloproteinase matrilysin (knockoutyMinycolon canceryextracellular matrixyAPC) CAROLE L. WILSON*†,KATHLEEN J. HEPPNER*†,PATRICIA A. LABOSKY*‡,BRIGID L. M. HOGAN*‡, AND LYNN M. MATRISIAN*§ *Department of Cell Biology and ‡Howard Hughes Medical Institute, Vanderbilt University, Nashville, TN 37232 Communicated by Ruth Sager, Dana–Farber Cancer Institute, Boston, MA, December 12, 1996 (received for review June 27, 1996) ABSTRACT Matrix metalloproteinases (MMPs) classi- The Min (multiple intestinal neoplasia) mouse has proven to cally have been implicated in basement membrane destruction be a powerful model system to study molecules involved in the associated with late-stage tumor cell invasion and metastasis. progression of intestinal adenomas. It has been determined However, recent studies have demonstrated that one MMP that a nonsense autosomal dominant germline mutation in the family member, matrilysin, is expressed in a high percentage adenomatous polyposis coli (Apc) gene (the ApcMin allele) of early-stage human colorectal tumors. We analyzed matri- induces spontaneous intestinal tumors in these mice, providing lysin expression in benign intestinal tumors from mice het- a model that closely mimics the human hereditary colon cancer erozygous for the ApcMin allele (Miny1) and found that the syndrome, familial adenomatous polyposis (13, 14). Congenic mRNA was induced in the majority (88%) of these adenomas. C57BLy6-Min mice heterozygous for this mutant allele Protein was detected in the tumor cells, where, surprisingly, (Miny1) develop numerous benign tumors throughout the it was predominantly immunolocalized to the lumenal surface intestinal tract, although with less frequency in the large bowel, of dysplastic glands rather than the basement membrane or and have an average life span of 119 days (13, 15). -

81969413.Pdf
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector European Journal of Pharmacology 698 (2013) 74–86 Contents lists available at SciVerse ScienceDirect European Journal of Pharmacology journal homepage: www.elsevier.com/locate/ejphar Molecular and cellular pharmacology Dipeptidyl peptidase IV inhibition upregulates GLUT4 translocation and expression in heart and skeletal muscle of spontaneously hypertensive rats Gisele Giannocco a,b, Kelen C. Oliveira a,b, Renato O. Crajoinas c, Gabriela Venturini c, Thiago A. Salles c, Miriam H. Fonseca-Alaniz c, Rui M.B. Maciel b, Adriana C.C. Girardi c,n a Department of Morphology and Physiology, Faculdade de Medicina do ABC, Santo Andre´,Sao~ Paulo, SP, Brazil b Department of Medicine, Federal University of Sao~ Paulo, Sao~ Paulo, SP, Brazil c University of Sao~ Paulo Medical School, Heart Institute, Laboratory of Genetics and Molecular Cardiology, Avenida Dr. Ene´as de Carvalho Aguiar 44, 101 andar, Bloco II, 05403-900 Sao Paulo, Brazil article info abstract Article history: The purpose of the current study was to test the hypothesis that the dipeptidyl peptidase IV (DPPIV) Received 26 January 2012 inhibitor sitagliptin, which exerts anti-hyperglycemic and anti-hypertensive effects, upregulates GLUT4 Received in revised form translocation, protein levels, and/or mRNA expression in heart and skeletal muscle of spontaneously 19 September 2012 hypertensive rats (SHRs). Ten days of treatment with sitagliptin (40 mg/kg twice daily) decreased Accepted 21 September 2012 plasma DPPIV activity in both young (Y, 5-week-old) and adult (A, 20-week-old) SHRs to similar Available online 7 October 2012 extents (85%). -

Discovery and Optimization of Selective Inhibitors of Meprin Α (Part II)
pharmaceuticals Article Discovery and Optimization of Selective Inhibitors of Meprin α (Part II) Chao Wang 1,2, Juan Diez 3, Hajeung Park 1, Christoph Becker-Pauly 4 , Gregg B. Fields 5 , Timothy P. Spicer 1,6, Louis D. Scampavia 1,6, Dmitriy Minond 2,7 and Thomas D. Bannister 1,2,* 1 Department of Molecular Medicine, Scripps Research, Jupiter, FL 33458, USA; [email protected] (C.W.); [email protected] (H.P.); [email protected] (T.P.S.); [email protected] (L.D.S.) 2 Department of Chemistry, Scripps Research, Jupiter, FL 33458, USA; [email protected] 3 Rumbaugh-Goodwin Institute for Cancer Research, Nova Southeastern University, 3321 College Avenue, CCR r.605, Fort Lauderdale, FL 33314, USA; [email protected] 4 The Scripps Research Molecular Screening Center, Scripps Research, Jupiter, FL 33458, USA; [email protected] 5 Unit for Degradomics of the Protease Web, Institute of Biochemistry, University of Kiel, Rudolf-Höber-Str.1, 24118 Kiel, Germany; fi[email protected] 6 Department of Chemistry & Biochemistry and I-HEALTH, Florida Atlantic University, 5353 Parkside Drive, Jupiter, FL 33458, USA 7 Dr. Kiran C. Patel College of Allopathic Medicine, Nova Southeastern University, 3301 College Avenue, Fort Lauderdale, FL 33314, USA * Correspondence: [email protected] Abstract: Meprin α is a zinc metalloproteinase (metzincin) that has been implicated in multiple diseases, including fibrosis and cancers. It has proven difficult to find small molecules that are capable Citation: Wang, C.; Diez, J.; Park, H.; of selectively inhibiting meprin α, or its close relative meprin β, over numerous other metzincins Becker-Pauly, C.; Fields, G.B.; Spicer, which, if inhibited, would elicit unwanted effects. -

2335 Roles of Molecules Involved in Epithelial/Mesenchymal Transition
[Frontiers in Bioscience 13, 2335-2355, January 1, 2008] Roles of molecules involved in epithelial/mesenchymal transition during angiogenesis Giulio Ghersi Dipartimento di Biologia Cellulare e dello Sviluppo, Universita di Palermo, Italy TABLE OF CONTENTS 1. Abstract 2. Introduction 3. Extracellular matrix 3.1. ECM and integrins 3.2. Basal lamina components 4. Cadherins. 4.1. Cadherins in angiogenesis 5. Integrins. 5.1. Integrins in angiogenesis 6. Focal adhesion molecules 7. Proteolytic enzymes 7.1. Proteolytic enzymes inhibitors 7.2. Proteolytic enzymes in angiogenesis 8. Perspective 9. Acknowledgements 10. References 1.ABSTRACT 2. INTRODUCTION Formation of vessels requires “epithelial- Growth of new blood vessels (angiogenesis) mesenchymal” transition of endothelial cells, with several plays a key role in several physiological processes, such modifications at the level of endothelial cell plasma as vascular remodeling during embryogenesis and membranes. These processes are associated with wound healing tissue repair in the adult; as well as redistribution of cell-cell and cell-substrate adhesion pathological processes, including rheumatoid arthritis, molecules, cross talk between external ECM and internal diabetic retinopathy, psoriasis, hemangiomas, and cytoskeleton through focal adhesion molecules and the cancer (1). Vessel formation entails the “epithelial- expression of several proteolytic enzymes, including matrix mesenchymal” transition of endothelial cells (ECs) “in metalloproteases and serine proteases. These enzymes with vivo”; a similar phenotypic exchange can be induced “in their degradative action on ECM components, generate vitro” by growing ECs to low cell density, or in “wound molecules acting as activators and/or inhibitors of healing” experiments or perturbing cell adhesion and angiogenesis. The purpose of this review is to provide an associated molecule functions. -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

3 Cleavage Products of Notch 2/Site and Myelopoiesis by Dysregulating
ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch This information is current as David R. Gibb, Sheinei J. Saleem, Dae-Joong Kang, Mark of October 4, 2021. A. Subler and Daniel H. Conrad J Immunol 2011; 186:4244-4252; Prepublished online 2 March 2011; doi: 10.4049/jimmunol.1003318 http://www.jimmunol.org/content/186/7/4244 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2011/03/02/jimmunol.100331 Material 8.DC1 http://www.jimmunol.org/ References This article cites 45 articles, 16 of which you can access for free at: http://www.jimmunol.org/content/186/7/4244.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 4, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch David R. -

ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease
International Journal of Molecular Sciences Review ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease Francesca Tosetti 1,* , Massimo Alessio 2, Alessandro Poggi 1,† and Maria Raffaella Zocchi 3,† 1 Molecular Oncology and Angiogenesis Unit, IRCCS Ospedale Policlinico S. Martino Largo R. Benzi 10, 16132 Genoa, Italy; [email protected] 2 Proteome Biochemistry, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] 3 Division of Immunology, Transplants and Infectious Diseases, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] * Correspondence: [email protected] † These authors contributed equally to this work as last author. Abstract: Enzymes, once considered static molecular machines acting in defined spatial patterns and sites of action, move to different intra- and extracellular locations, changing their function. This topological regulation revealed a close cross-talk between proteases and signaling events involving post-translational modifications, membrane tyrosine kinase receptors and G-protein coupled recep- tors, motor proteins shuttling cargos in intracellular vesicles, and small-molecule messengers. Here, we highlight recent advances in our knowledge of regulation and function of A Disintegrin And Metalloproteinase (ADAM) endopeptidases at specific subcellular sites, or in multimolecular com- plexes, with a special focus on ADAM10, and tumor necrosis factor-α convertase (TACE/ADAM17), since these two enzymes belong to the same family, share selected substrates and bioactivity. We will discuss some examples of ADAM10 activity modulated by changing partners and subcellular compartmentalization, with the underlying hypothesis that restraining protease activity by spatial Citation: Tosetti, F.; Alessio, M.; segregation is a complex and powerful regulatory tool. -

Cell-Autonomous FLT3L Shedding Via ADAM10 Mediates Conventional Dendritic Cell Development in Mouse Spleen
Cell-autonomous FLT3L shedding via ADAM10 mediates conventional dendritic cell development in mouse spleen Kohei Fujitaa,b,1, Svetoslav Chakarovc,1, Tetsuro Kobayashid, Keiko Sakamotod, Benjamin Voisind, Kaibo Duanc, Taneaki Nakagawaa, Keisuke Horiuchie, Masayuki Amagaib, Florent Ginhouxc, and Keisuke Nagaod,2 aDepartment of Dentistry and Oral Surgery, Keio University School of Medicine, Tokyo 160-8582, Japan; bDepartment of Dermatology, Keio University School of Medicine, Tokyo 160-8582, Japan; cSingapore Immunology Network, Agency for Science, Technology and Research, Biopolis, 138648 Singapore; dDermatology Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892; and eDepartment of Orthopedic Surgery, National Defense Medical College, Tokorozawa 359-8513, Japan Edited by Kenneth M. Murphy, Washington University School of Medicine, St. Louis, MO, and approved June 10, 2019 (received for review November 4, 2018) Conventional dendritic cells (cDCs) derive from bone marrow (BM) intocDC1sorcDC2stakesplaceintheBM(3),andthese precursors that undergo cascades of developmental programs to pre-cDC1s and pre-cDC2s ultimately differentiate into cDC1s terminally differentiate in peripheral tissues. Pre-cDC1s and pre- and cDC2s after migrating to nonlymphoid and lymphoid tissues. + + cDC2s commit in the BM to each differentiate into CD8α /CD103 cDCs are short-lived, and their homeostatic maintenance relies + cDC1s and CD11b cDC2s, respectively. Although both cDCs rely on on constant replenishment from the BM precursors (5). The cy- the cytokine FLT3L during development, mechanisms that ensure tokine Fms-related tyrosine kinase 3 ligand (FLT3L) (12), by cDC accessibility to FLT3L have yet to be elucidated. Here, we gen- signaling through its receptor FLT3 expressed on DC precursors, erated mice that lacked a disintegrin and metalloproteinase (ADAM) is essential during the development of DCs (7, 13). -
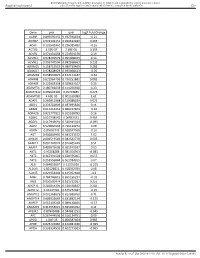
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al.