Microrna Expression Profiling of Peripheral Blood Mononuclear Cells Associated with Syphilis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Two Freshwater Shrimp Species of the Genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the Description of a New Species
A peer-reviewed open-access journal ZooKeys 923: 15–32 (2020) Caridina tetrazona 15 doi: 10.3897/zookeys.923.48593 RESEarcH articLE http://zookeys.pensoft.net Launched to accelerate biodiversity research Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species Qing-Hua Chen1, Wen-Jian Chen2, Xiao-Zhuang Zheng2, Zhao-Liang Guo2 1 South China Institute of Environmental Sciences, Ministry of Ecology and Environment, Guangzhou 510520, Guangdong Province, China 2 Department of Animal Science, School of Life Science and Enginee- ring, Foshan University, Foshan 528231, Guangdong Province, China Corresponding author: Zhao-Liang Guo ([email protected]) Academic editor: I.S. Wehrtmann | Received 19 November 2019 | Accepted 7 February 2020 | Published 1 April 2020 http://zoobank.org/138A88CC-DF41-437A-BA1A-CB93E3E36D62 Citation: Chen Q-H, Chen W-J, Zheng X-Z, Guo Z-L (2020) Two freshwater shrimp species of the genus Caridina (Decapoda, Caridea, Atyidae) from Dawanshan Island, Guangdong, China, with the description of a new species. ZooKeys 923: 15–32. https://doi.org/10.3897/zookeys.923.48593 Abstract A faunistic and ecological survey was conducted to document the diversity of freshwater atyid shrimps of Dawanshan Island. Two species of Caridina that occur on this island were documented and discussed. One of these, Caridina tetrazona sp. nov. is described and illustrated as new to science. It can be easily distinguished from its congeners based on a combination of characters, which includes a short rostrum, the shape of the endopod of the male first pleopod, the segmental ratios of antennular peduncle and third maxilliped, the slender scaphocerite, and the absence of a median projection on the posterior margin. -

EICC-Gesi Conflict Minerals Reporting Template
The following list represents the CFSI's latest smelter name/alias information as of this templates release. This list is updated frequently, and the most up-to-date version can be found on the CFSI website http://www.conflictfreesourcing.org/conflict-free-smelter-program/exports/cmrt-export/. The presence of a smelter here is NOT a guarantee that it is currently Active or Compliant within the Conflict-Free Smelter Program. Please refer to the CFSI web site www.conflictfreesourcing.org for the most current and accurate list of standard smelter names that are Active or Compliant. Names included in column B represent company names that are commonly recognized and reported by the supply chain for a particular smelter. These names may include former company names, alternate names, abbreviations, or other variations. Although the names may not be the CFSI Standard Smelter Name, the reference names are helpful to identify the smelter, which is listed under column C in the Smelter Reference List. Column C is the list of the official standard smelter names, understood to be the legal names of the eligible smelters. The majority of smelters will have the same entry for both columns, however Metal Smelter Reference List Gold Abington Reldan Metals, LLC Gold Accurate Refining Group Gold Advanced Chemical Company Gold AGR Mathey Gold AGR(Perth Mint Australia) Gold Aida Chemical Industries Co., Ltd. Gold Al Etihad Gold Refinery DMCC Gold Allgemeine Gold-und Silberscheideanstalt A.G. Gold Almalyk Mining and Metallurgical Complex (AMMC) Gold Amagasaki Factory, Hyogo Prefecture, Japan Gold AngloGold Ashanti Córrego do Sítio Mineração Gold Anhui Tongling Nonferrous Metal Mining Co., Ltd. -

Guangdong Province, 2019
China CDC Weekly Preplanned Studies Co-Administration of Multiple Childhood Vaccines — Guangdong Province, 2019 Hai Li1,2; Yanqiu Tan1,3; Haiying Zeng1,3; Fengmei Zeng1,4; Xing Xu1,5; Yu Liao1,6; Qi Zhu6; Meng Zhang1,6; Xuguang Chen1,6; Min Kang1,6; Fujie Xu7; Huizhen Zheng1,6,# This policy could save about 1137.62 RMB for each Summary child during their first 2 years of life. To provide scope, What is already known about this topic? 1.8 million infants in Guangdong received the first The Co-Administration of Multiple Vaccines were dose of Hepatitis B vaccine in 2018; based on the implemented in many countries and have been shown number of children, this policy could therefore save up to significantly reduce many times of visiting the to 2.0 billion RMB for families in Guangdong vaccination clinic. Province for this single vaccination event. The Co- What is added by this report? Administration of Multiple Vaccines Policy can It is the first time to calculate the cost of visiting significantly reduce vaccination costs for children’s vaccination clinic from transportation and work- families and can greatly improve the social cost- effectiveness of childhood vaccinations. Our findings absence for children’s families in Guangdong. suggest that Co-Administration of Multiple Vaccines What are the implications for public health should be implemented as soon as possible. practice? This study estimated the cost incurred by the We demonstrated the importance of Co- families with children under 2 years old in Guangdong Administration of Multiple Vaccines that reduce the Province during the process of inoculation. -

Shop Direct Factory List Dec 18
Factory Factory Address Country Sector FTE No. workers % Male % Female ESSENTIAL CLOTHING LTD Akulichala, Sakashhor, Maddha Para, Kaliakor, Gazipur, Bangladesh BANGLADESH Garments 669 55% 45% NANTONG AIKE GARMENTS COMPANY LTD Group 14, Huanchi Village, Jiangan Town, Rugao City, Jaingsu Province, China CHINA Garments 159 22% 78% DEEKAY KNITWEARS LTD SF No. 229, Karaipudhur, Arulpuram, Palladam Road, Tirupur, 641605, Tamil Nadu, India INDIA Garments 129 57% 43% HD4U No. 8, Yijiang Road, Lianhang Economic Development Zone, Haining CHINA Home Textiles 98 45% 55% AIRSPRUNG BEDS LTD Canal Road, Canal Road Industrial Estate, Trowbridge, Wiltshire, BA14 8RQ, United Kingdom UK Furniture 398 83% 17% ASIAN LEATHERS LIMITED Asian House, E. M. Bypass, Kasba, Kolkata, 700017, India INDIA Accessories 978 77% 23% AMAN KNITTINGS LIMITED Nazimnagar, Hemayetpur, Savar, Dhaka, Bangladesh BANGLADESH Garments 1708 60% 30% V K FASHION LTD formerly STYLEWISE LTD Unit 5, 99 Bridge Road, Leicester, LE5 3LD, United Kingdom UK Garments 51 43% 57% AMAN GRAPHIC & DESIGN LTD. Najim Nagar, Hemayetpur, Savar, Dhaka, Bangladesh BANGLADESH Garments 3260 40% 60% WENZHOU SUNRISE INDUSTRIAL CO., LTD. Floor 2, 1 Building Qiangqiang Group, Shanghui Industrial Zone, Louqiao Street, Ouhai, Wenzhou, Zhejiang Province, China CHINA Accessories 716 58% 42% AMAZING EXPORTS CORPORATION - UNIT I Sf No. 105, Valayankadu, P. Vadugapal Ayam Post, Dharapuram Road, Palladam, 541664, India INDIA Garments 490 53% 47% ANDRA JEWELS LTD 7 Clive Avenue, Hastings, East Sussex, TN35 5LD, United Kingdom UK Accessories 68 CAVENDISH UPHOLSTERY LIMITED Mayfield Mill, Briercliffe Road, Chorley Lancashire PR6 0DA, United Kingdom UK Furniture 33 66% 34% FUZHOU BEST ART & CRAFTS CO., LTD No. 3 Building, Lifu Plastic, Nanshanyang Industrial Zone, Baisha Town, Minhou, Fuzhou, China CHINA Homewares 44 41% 59% HUAHONG HOLDING GROUP No. -

20200316 Factory List.Xlsx
Country Factory Name Address BANGLADESH AMAN WINTER WEARS LTD. SINGAIR ROAD, HEMAYETPUR, SAVAR, DHAKA.,0,DHAKA,0,BANGLADESH BANGLADESH KDS GARMENTS IND. LTD. 255, NASIRABAD I/A, BAIZID BOSTAMI ROAD,,,CHITTAGONG-4211,,BANGLADESH BANGLADESH DENITEX LIMITED 9/1,KORNOPARA, SAVAR, DHAKA-1340,,DHAKA,,BANGLADESH JAMIRDIA, DUBALIAPARA, VALUKA, MYMENSHINGH BANGLADESH PIONEER KNITWEARS (BD) LTD 2240,,MYMENSHINGH,DHAKA,BANGLADESH PLOT # 49-52, SECTOR # 08 , CEPZ, CHITTAGONG, BANGLADESH HKD INTERNATIONAL (CEPZ) LTD BANGLADESH,,CHITTAGONG,,BANGLADESH BANGLADESH FLAXEN DRESS MAKER LTD MEGHDUBI, WARD: 40, GAZIPUR CITY CORP,,,GAZIPUR,,BANGLADESH BANGLADESH NETWORK CLOTHING LTD 228/3,SHAHID RAWSHAN SARAK, CHANDANA,,,GAZIPUR,DHAKA,BANGLADESH 521/1 GACHA ROAD, BOROBARI,GAZIPUR CITY BANGLADESH ABA FASHIONS LTD CORPORATION,,GAZIPUR,DHAKA,BANGLADESH VILLAGE- AMTOIL, P.O. HAT AMTOIL, P.S. SREEPUR, DISTRICT- BANGLADESH SAN APPARELS LTD MAGURA,,JESSORE,,BANGLADESH BANGLADESH TASNIAH FABRICS LTD KASHIMPUR NAYAPARA, GAZIPUR SADAR,,GAZIPUR,,BANGLADESH BANGLADESH AMAN KNITTINGS LTD KULASHUR, HEMAYETPUR,,SAVAR,DHAKA,BANGLADESH BANGLADESH CHERRY INTIMATE LTD PLOT # 105 01,DEPZ, ASHULIA, SAVAR,DHAKA,DHAKA,BANGLADESH COLOMESSHOR, POST OFFICE-NATIONAL UNIVERSITY, GAZIPUR BANGLADESH ARRIVAL FASHION LTD SADAR,,,GAZIPUR,DHAKA,BANGLADESH VILLAGE-JOYPURA, UNION-SHOMBAG,,UPAZILA-DHAMRAI, BANGLADESH NAFA APPARELS LTD DISTRICT,DHAKA,,BANGLADESH BANGLADESH VINTAGE DENIM APPARELS LIMITED BOHERARCHALA , SREEPUR,,,GAZIPUR,,BANGLADESH BANGLADESH KDS IDR LTD CDA PLOT NO: 15(P),16,MOHORA -

The Stoor Hobbit of Guangdong: Goniurosaurus Gollum Sp. Nov., a Cave-Dwelling Leopard Gecko (Squamata, Eublepharidae) from South China
ZooKeys 991: 137–153 (2020) A peer-reviewed open-access journal doi: 10.3897/zookeys.991.54935 RESEARCH ARTICLE https://zookeys.pensoft.net Launched to accelerate biodiversity research The Stoor Hobbit of Guangdong: Goniurosaurus gollum sp. nov., a cave-dwelling Leopard Gecko (Squamata, Eublepharidae) from South China Shuo Qi1,*, Jian Wang1,*, L. Lee Grismer2, Hong-Hui Chen1, Zhi-Tong Lyu1, Ying-Yong Wang1 1 State Key Laboratory of Biocontrol/ The Museum of Biology, School of Life Sciences, Sun Yat-sen University, Guangzhou, Guangdong 510275, China 2 Herpetology Laboratory, Department of Biology, La Sierra Univer- sity, Riverside, California 92515, USA Corresponding author: Ying-Yong Wang ([email protected]) Academic editor: T. Ziegler | Received 31 May 2020 | Accepted 10 September 2020 | Published 11 November 2020 http://zoobank.org/2D9EEFC0-B43E-4AC3-86E7-89944E54169B Citation: Qi S, Wang J, Grismer LL, Chen H-H, Lyu Z-T, Wang Y-Y (2020) The Stoor Hobbit of Guangdong: Goniurosaurus gollum sp. nov., a cave-dwelling Leopard Gecko (Squamata, Eublepharidae) from South China. ZooKeys 991: 137–153. https://doi.org/10.3897/zookeys.991.54935 Abstract A new species of the genus Goniurosaurus is described based on three specimens collected from a limestone cave in Huaiji County, Guangdong Province, China. Based on molecular phylogenetic analyses, the new species is nested within the Goniurosaurus yingdeensis species group. However, morphological analyses cannot ascribe it to any known species of that group. It is distinguished from the other three species in the group by a combination of the following characters: scales around midbody 121–128; dorsal tubercle rows at midbody 16–17; presence of 10–11 precloacal pores in males, and absent in females; nuchal loop and body bands immaculate, without black spots; iris orange, gradually darker on both sides. -
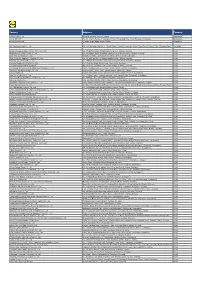
Factory Address Country
Factory Address Country Durable Plastic Ltd. Mulgaon, Kaligonj, Gazipur, Dhaka Bangladesh Lhotse (BD) Ltd. Plot No. 60&61, Sector -3, Karnaphuli Export Processing Zone, North Potenga, Chittagong Bangladesh Bengal Plastics Ltd. Yearpur, Zirabo Bazar, Savar, Dhaka Bangladesh ASF Sporting Goods Co., Ltd. Km 38.5, National Road No. 3, Thlork Village, Chonrok Commune, Korng Pisey District, Konrrg Pisey, Kampong Speu Cambodia Ningbo Zhongyuan Alljoy Fishing Tackle Co., Ltd. No. 416 Binhai Road, Hangzhou Bay New Zone, Ningbo, Zhejiang China Ningbo Energy Power Tools Co., Ltd. No. 50 Dongbei Road, Dongqiao Industrial Zone, Haishu District, Ningbo, Zhejiang China Junhe Pumps Holding Co., Ltd. Wanzhong Villiage, Jishigang Town, Haishu District, Ningbo, Zhejiang China Skybest Electric Appliance (Suzhou) Co., Ltd. No. 18 Hua Hong Street, Suzhou Industrial Park, Suzhou, Jiangsu China Zhejiang Safun Industrial Co., Ltd. No. 7 Mingyuannan Road, Economic Development Zone, Yongkang, Zhejiang China Zhejiang Dingxin Arts&Crafts Co., Ltd. No. 21 Linxian Road, Baishuiyang Town, Linhai, Zhejiang China Zhejiang Natural Outdoor Goods Inc. Xiacao Village, Pingqiao Town, Tiantai County, Taizhou, Zhejiang China Guangdong Xinbao Electrical Appliances Holdings Co., Ltd. South Zhenghe Road, Leliu Town, Shunde District, Foshan, Guangdong China Yangzhou Juli Sports Articles Co., Ltd. Fudong Village, Xiaoji Town, Jiangdu District, Yangzhou, Jiangsu China Eyarn Lighting Ltd. Yaying Gang, Shixi Village, Shishan Town, Nanhai District, Foshan, Guangdong China Lipan Gift & Lighting Co., Ltd. No. 2 Guliao Road 3, Science Industrial Zone, Tangxia Town, Dongguan, Guangdong China Zhan Jiang Kang Nian Rubber Product Co., Ltd. No. 85 Middle Shen Chuan Road, Zhanjiang, Guangdong China Ansen Electronics Co. Ning Tau Administrative District, Qiao Tau Zhen, Dongguan, Guangdong China Changshu Tongrun Auto Accessory Co., Ltd. -

Cassava in China Inad• Era of Change
, '. -.:. " . Ie'"d;~~aVa in China lnan• I j Era of Change A CBN Case Study with Farmers and Processors ~-- " '. -.-,'" . ,; . ):.'~. - ...~. ¡.;; i:;f;~ ~ ';. ~:;':. __ ~~,.:';.: GuyHenry an~ Reinhardt Howeler )28103 U.' '1'/ "'.'..,· •.. :¡g.l ... !' . ~ .. W()R~mG,~6t:UMENT 1§:º~~U'U~T'O~OIln1ernotlonol CeMe:r fer TropIcal AgrICultura No. 155 Cassava Biotechnolgy Network Cassava in China InaD• Era of Change A CBN Case Study with Farmers and Processors GuyHenry and Reinhardt Howeler Cover Photos: Top: Cassava processing in Southern China í Bottom: Farmer participatory research in Southern China I I Al! photos: Cuy Henry (ClAn, July-August, 1994 I I¡ ¡ ¡, I Centro Internacional de Agricultura Tropical, CIAT ! Intemational Center for Tropical Agriculwre I Apartado Aéreo 6713 Cali, Colombia G:IAT Working Document No. 155 Press fun: 100 Printed in Colombia june 1996 ! Correa citation: Henry, G.; Howeler, R. 1996. Cassava in China in an era of change. A CBN case study with farmers and processors. 31 July to 20 August, 1994. - Cali Colombia: Centro Internacional de Agricultura Tropical, 1996. 68 p. - (Working Document; no. 1 ~5) I Cassava in China in An Era of Change A CBN Case Study with farmers and processors in Guangdong, Guangxi and Hainan Provinces of Southern China By: Guy Henry and Reínhardt Howeler luly 31 - August 20, 1994 Case Study Team Members: Dr. Guy Henry (Economist) International Center for Tropical Agriculture (ClAn, Cal i, Colombia Dr. Reinharot Howeler (Agronomis!) Intemational Center for Tropical Agricultur<! (ClAn, Bangkok, Thailand Mr. Huang Hong Cheng (Director), Mr. Fang Baiping, M •. Fu Guo Hui 01 the Upland Crops Researcll Institute (UCRIl in Guangzhou. -

Sites of Zou and Ma Migration in the Qing
APPENDIX F Sites of Zou and Ma Migration in the Qing The information on Zou and Ma migration is drawn from the generational charts of the three primary genealogies, the MSDZZP (1945), FYZSZP (1947), and MTLZXZSZP (1911). Since precise dates of migration are given only very rarely, I have estimated the probable time of migration from the birth and death dates given. “Zou1” refers to the upper-shrine Zou lineage; “Zou2” to the lower-shrine. The table is organized by province, prefecture, and county. ________________________________________________________________________________________________ Late 17th– Late 18th– Late 19th– early 18th mid-19th early 20th Bookselling Location century century century site ________________________________________________________________________________________________ Anhui province Wuyuan 婺源 county (site of Ma migration in the late 16th c.) Guangdong province Chaozhou 潮州 prefecture Ma, Zou1 Zou2 * Chenghai 澄海 county Ma Dabu 大埔 county Ma, Zou2 * Jieyang 揭陽 county Ma * Guangzhou 廣州 prefecture Zou2 * Foshanzhen 佛山鎮 Zou1 * Qingyuan 清遠 county Zou2 Gaozhou 高州 prefecture Ma, Zou1 * Dianbai 電白 county Zou1 Huazhou 化州 county Zou1 Xinyi 信宜 county Ma Huizhou 惠州 prefecture Ma Zou2 * Boluo 博羅 county Zou2 Changning 長寧 county Ma Haifeng 海豐 county Ma Heping 和平 county (a site of Ma migration in the late 16th c.) Ma * Heyuan 河源 county Ma, Zou2 * Kuzhupai 苦竹派 village or market Ma, Zou1 Lianpingzhou 連平州 county Ma, Zou2 Longchuan 龍川 county Zou1, Zou2 * Yongan 永安 county Zou1 Zou1 * Jiayingzhou 嘉應 ( Jiaying department, a site of Zou1 migration in the late 16th c.) Zou2 Ma Ma * Changle 長樂 county Ma * Pingyuan 平遠 county Ma * Xingning 興寧 county Zou2 Ma * Zhenping 鎮平 county Ma, Zou1 Zou1 * Leizhou 雷州 prefecture Zou2 * Lianzhou 連州 prefecture Zou2 Zou2 Lianzhou 廉州 prefecture Zou1, Zou2 Hepu 合浦 county Zou1 Lingshan 靈山 county Ma, Zou1 Zou2 * Luodingzhou 羅定州 (Luoding department) Zou2 Nanxiong 南雄 prefecture Ma * Shixing 始興 county Ma Zou1, Zou2 * Qiongzhou 瓊州 prefecture Ma Zou1 Haikou 海口 city Zou1 Lingshui 陵水 county Zou2 Brokaw, Appendix F, p. -
![Directors, Supervisors and Parties Involved in the [Redacted]](https://docslib.b-cdn.net/cover/6866/directors-supervisors-and-parties-involved-in-the-redacted-1826866.webp)
Directors, Supervisors and Parties Involved in the [Redacted]
THIS DOCUMENT IS IN DRAFT FORM, INCOMPLETE AND SUBJECT TO CHANGE AND THAT THE INFORMATION MUST BE READ IN CONJUNCTION WITH THE SECTION HEADED “WARNING” ON THE COVER OF THIS DOCUMENT. DIRECTORS, SUPERVISORS AND PARTIES INVOLVED IN THE [REDACTED] DIRECTORS Name Residential Address Nationality Executive Directors Mr. Wang Jikang ( ) (Chairman) .................. Room 1101, Building J Chinese Central Courtyard Huijing North Street Favorview Palace Tianhe District Guangzhou Guangdong Province PRC Mr. Yi Xuefei ( ) (Vice Chairman) .............. 385-2, Yahuju Agile Garden Chinese 398 Xingnan Avenue Nancun Town Panyu District Guangzhou Guangdong Province PRC Mr. Wu Huiqiang ( ) ........ Room 1604 Chinese No. 38 Taojin Street Yuexiu District Guangzhou Guangdong Province PRC Non-executive Directors Mr. Su Zhigang ( ).......... No.18Zhupo Alley Chinese Gaodi Street Dashi Street Panyu District Guangzhou Guangdong Province PRC Mr. Shao Jianming ( ) ....... No.25Huayuan Yi Street Chinese Yuexiu District Guangzhou Guangdong Province PRC Mr. Li Fangjin ( )........... Room 203, Building 29 Chinese Middle District of South China Normal University No. 55 Zhongshan Road West Tianhe District Guangzhou Guangdong Province PRC 75 THIS DOCUMENT IS IN DRAFT FORM, INCOMPLETE AND SUBJECT TO CHANGE AND THAT THE INFORMATION MUST BE READ IN CONJUNCTION WITH THE SECTION HEADED “WARNING” ON THE COVER OF THIS DOCUMENT. DIRECTORS, SUPERVISORS AND PARTIES INVOLVED IN THE [REDACTED] Name Residential Address Nationality Mr. Zheng Shuping ( )....... Room 2301 Chinese No. 134 Huacheng Road Tianhe District Guangzhou Guangdong Province PRC Mr. Zhu Kelin ( ) ........... Room 2903 Chinese No. 38 Huajing North Street Tianhe District Guangzhou Guangdong Province PRC Mr. Zhang Yongming ( ) ..... 29-3-601 WanQuan Xinxin Jiayuan Chinese Wanliu East Road Haidian District Beijing PRC Mr. -

Industry Overview
THIS DOCUMENT IS IN DRAFT FORM, INCOMPLETE AND SUBJECT TO CHANGE AND THAT THE INFORMATION MUST BE READ IN CONJUNCTION WITH THE SECTION HEADED “WARNING” ON THE COVER OF THIS DOCUMENT. INDUSTRY OVERVIEW This section contains certain statistics, industry data or other information which have been derived from government, official or other public sources. We believe that the sources of such information are appropriate sources for such information and have taken reasonable care in extracting and reproducing such information. We have no reason to believe that such information is false or misleading or that any fact has been omitted that would render such information false or misleading. The information has not been independently verified by us, the Sole Sponsor, [REDACTED], the [REDACTED], any of their respective directors, officers, affiliates, advisors or representatives, or any other party involved in the [REDACTED], and no representation is given as to its accuracy. We, the Sole Sponsor, [REDACTED], any of their respective directors, officers, affiliates, advisors or representatives, and any other party involved in the [REDACTED] make no representation as to the completeness, accuracy or fairness of such information and accordingly such information should not be unduly relied upon. SOURCE OF INFORMATION The Company commissioned the C&W Report from Cushman & Wakefield (C&W) relating to the economy of the PRC, the residential property market in Guangdong, Hainan, Yunnan and Hunan provinces and the industries in which our Group operates. C&W has charged a total fee of HK$800,000 for the preparation of the C&W Report. C&W is a global real estate adviser, which offers a range of services including investment agency, leasing agency, property and facilities management, project and building consultancy, investment and asset management, market research and forecasting and valuation. -

2.11 Guangdong.Pdf
2.11 Guangdong Province Guangdong Guangyu Group Co., Ltd., affiliated to the Guangdong Provincial Prison Administration Bureau, has 26 prisons and one police hospital under its jurisdiction Legal representative of the prison company: Li Jingyan, Chairman of Guangdong Guangyu Group Co., Ltd. His official positions in the prison system: Communist Party Committee Member of Guangdong Provincial Department of Justice; Communist Party Committee Secretary and Director of Guangdong Provincial Prison Administration Bureau1 Guangdong Guangyu Group Co., Ltd. was built in July 1951, and it is located in Yujiao Town, Jiedong District, Jieyang City, Guangdong Province. It adopted its current name in May 1995. From 1993 to 2006, the prison fully implemented its “two strategic transfers” (which are the transformation from the field agricultural production to in-prison industrial production in terms of its industrial structure and the transfer from its previous remote mountainous area to the city in terms of its prison layout). The prison was moved from the remote mountainous area to its present location, which covers an area of more than 340 mu. Its prison production projects include electronic product processing, sewing, hardware processing, and manual processing. No. Company Name of the Legal Person Legal Registered Business Scope Company Notes on the Prison Name Prison, to and representative/ Capital Address which the Shareholder(s) Title Company Belongs 1 Guangdong Guangdong Guangdong Li Jingyan 863.874 Project investments, corporate Rooms 107, The Guangdong Provincial Prison Administration Guangyu Provincial Provincial Chairman, million yuan planning management. 302, 401, Bureau3 is the penalty enforcement department of the Group Co., Prison Prison Guangdong Building No.