Phyre 2 Results for O75828
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Redalyc.Microorganisms Screening for Limonene Oxidation
Ciência e Tecnologia de Alimentos ISSN: 0101-2061 [email protected] Sociedade Brasileira de Ciência e Tecnologia de Alimentos Brasil LERIN, Lindomar; TONIAZZO, Geciane; de OLIVEIRA, Débora; ROTTAVA, Leda; DARIVA, Cláudio; CANSIAN, Rogério Luis; TREICHEL, Helen; PADILHA, Francine; Ceva ANTUNES, Octávio Augusto Microorganisms screening for limonene oxidation Ciência e Tecnologia de Alimentos, vol. 30, núm. 2, abril-junio, 2010, pp. 399-405 Sociedade Brasileira de Ciência e Tecnologia de Alimentos Campinas, Brasil Available in: http://www.redalyc.org/articulo.oa?id=395940100017 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Ciência e Tecnologia de Alimentos ISSN 0101-2061 Microorganisms screening for limonene oxidation Seleção de microrganismos para oxidação de limoneno Original Lindomar LERIN1, Geciane TONIAZZO2, Débora de OLIVEIRA2*, Leda ROTTAVA1, Cláudio DARIVA3, Rogério Luis CANSIAN2, Helen TREICHEL2, Francine PADILHA3, Octávio Augusto Ceva ANTUNES1 Abstract Limonene is a monoterpene obtained in large amounts from essential oils and is used as a raw material for the synthesis of flavors and fine chemicals. Several pathways or routes for the microbial degradation of limonene making use of the cytochrome P450-dependent monooxygenases have been described. In this study, we present a fermentative screening of microorganisms in order to verify their ability to perform the desirable conversion. In parallel, the PCR technique was used to select the microorganisms that contain the limC gene, which is responsible for the conversion of carveol to carvone. -

Biosynthesis of New Alpha-Bisabolol Derivatives Through a Synthetic Biology Approach Arthur Sarrade-Loucheur
Biosynthesis of new alpha-bisabolol derivatives through a synthetic biology approach Arthur Sarrade-Loucheur To cite this version: Arthur Sarrade-Loucheur. Biosynthesis of new alpha-bisabolol derivatives through a synthetic biology approach. Biochemistry, Molecular Biology. INSA de Toulouse, 2020. English. NNT : 2020ISAT0003. tel-02976811 HAL Id: tel-02976811 https://tel.archives-ouvertes.fr/tel-02976811 Submitted on 23 Oct 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. THÈSE En vue de l’obtention du DOCTORAT DE L’UNIVERSITÉ DE TOULOUSE Délivré par l'Institut National des Sciences Appliquées de Toulouse Présentée et soutenue par Arthur SARRADE-LOUCHEUR Le 30 juin 2020 Biosynthèse de nouveaux dérivés de l'α-bisabolol par une approche de biologie synthèse Ecole doctorale : SEVAB - Sciences Ecologiques, Vétérinaires, Agronomiques et Bioingenieries Spécialité : Ingénieries microbienne et enzymatique Unité de recherche : TBI - Toulouse Biotechnology Institute, Bio & Chemical Engineering Thèse dirigée par Gilles TRUAN et Magali REMAUD-SIMEON Jury -

Solarbio Catalogue with PRICES
CAS Name Grade Purity Biochemical Reagent Biochemical Reagent 75621-03-3 C8390-1 3-((3-Cholamidopropyl)dimethylammonium)-1-propanesulfonateCHAPS Ultra Pure Grade 1g 75621-03-3 C8390-5 3-((3-Cholamidopropyl)dimethylammonium)-1-propanesulfonateCHAPS 5g 57-09-0 C8440-25 Cetyl-trimethyl Ammonium Bromide CTAB High Pure Grade ≥99.0% 25g 57-09-0 C8440-100 Cetyl-trimethyl Ammonium Bromide CTAB High Pure Grade ≥99.0% 100g 57-09-0 C8440-500 Cetyl-trimethyl Ammonium Bromide CTAB High Pure Grade ≥99.0% 500g E1170-100 0.5M EDTA (PH8.0) 100ml E1170-500 0.5M EDTA (PH8.0) 500ml 6381-92-6 E8030-100 EDTA disodium salt dihydrate EDTA Na2 Biotechnology Grade ≥99.0% 100g 6381-92-6 E8030-500 EDTA disodium salt dihydrate EDTA Na2 Biotechnology Grade ≥99.0% 500g 6381-92-6 E8030-1000 EDTA disodium salt dihydrate EDTA Na2 Biotechnology Grade ≥99.0% 1kg 6381-92-6 E8030-5000 EDTA disodium salt dihydrate EDTA Na2 Biotechnology Grade ≥99.0% 5kg 60-00-4 E8040-100 Ethylenediaminetetraacetic acid EDTA Ultra Pure Grade ≥99.5% 100g 60-00-4 E8040-500 Ethylenediaminetetraacetic acid EDTA Ultra Pure Grade ≥99.5% 500g 60-00-4 E8040-1000 Ethylenediaminetetraacetic acid EDTA Ultra Pure Grade ≥99.5% 1kg 67-42-5 E8050-5 Ethylene glycol-bis(2-aminoethylether)-N,N,NEGTA′,N′-tetraacetic acid Ultra Pure Grade ≥97.0% 5g 67-42-5 E8050-10 Ethylene glycol-bis(2-aminoethylether)-N,N,NEGTA′,N′-tetraacetic acid Ultra Pure Grade ≥97.0% 10g 50-01-1 G8070-100 Guanidine Hydrochloride Guanidine HCl ≥98.0%(AT) 100g 50-01-1 G8070-500 Guanidine Hydrochloride Guanidine HCl ≥98.0%(AT) 500g 56-81-5 -

Fungal Pathogenesis in Humans the Growing Threat
Fungal Pathogenesis in Humans The Growing Threat Edited by Fernando Leal Printed Edition of the Special Issue Published in Genes www.mdpi.com/journal/genes Fungal Pathogenesis in Humans Fungal Pathogenesis in Humans The Growing Threat Special Issue Editor Fernando Leal MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade Special Issue Editor Fernando Leal Instituto de Biolog´ıa Funcional y Genomica/Universidad´ de Salamanca Spain Editorial Office MDPI St. Alban-Anlage 66 4052 Basel, Switzerland This is a reprint of articles from the Special Issue published online in the open access journal Genes (ISSN 2073-4425) from 2018 to 2019 (available at: https://www.mdpi.com/journal/genes/special issues/Fungal Pathogenesis Humans Growing Threat). For citation purposes, cite each article independently as indicated on the article page online and as indicated below: LastName, A.A.; LastName, B.B.; LastName, C.C. Article Title. Journal Name Year, Article Number, Page Range. ISBN 978-3-03897-900-5 (Pbk) ISBN 978-3-03897-901-2 (PDF) Cover image courtesy of Fernando Leal. c 2019 by the authors. Articles in this book are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book as a whole is distributed by MDPI under the terms and conditions of the Creative Commons license CC BY-NC-ND. Contents About the Special Issue Editor ...................................... vii Fernando Leal Special Issue: Fungal Pathogenesis in Humans: The Growing Threat Reprinted from: Genes 2019, 10, 136, doi:10.3390/genes10020136 .................. -

Investigating Labdane-Related Diterpenoids Biosynthetic Network In
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2012 Investigating labdane-related diterpenoids biosynthetic network in rice: characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase Yisheng Wu Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Biochemistry Commons Recommended Citation Wu, Yisheng, "Investigating labdane-related diterpenoids biosynthetic network in rice: characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase" (2012). Graduate Theses and Dissertations. 12680. https://lib.dr.iastate.edu/etd/12680 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Investigating labdane-related diterpenoids biosynthetic network in rice: Characterization of cytochromes P450 and short-chain alcohol dehydrogenases/reductase by Yisheng Wu A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Biochemistry Program of Study Committee: Reuben Peters, Major Professor Gustavo MacIntosh Thomas Bobik Bing Yang Mark Hargrove Iowa State University Ames, Iowa 2012 Copyright © Yisheng Wu , 2012. All -

Pea SAD Short-Chain Dehydrogenase/Reductase Author to Whom All Correspondence Should Be Addressed
Plant Physiology Preview. Published on February 22, 2011, as DOI:10.1104/pp.111.173336 1 Running head: Pea SAD Short-Chain Dehydrogenase/Reductase Author to whom all correspondence should be addressed: Åke Strid Akademin för naturvetenskap och teknik och Centrum för livsvetenskap Örebro universitet SE-70182 Örebro Sweden Telephone: +46-19-303603 E-mail: [email protected] Most appropriate journal research area: Cell Biology 1 Copyright 2011 by the American Society of Plant Biologists 2 The Pisum sativum SAD Short-Chain Dehydrogenase/Reductase: Quinone Reduction, Tissue 1 Distribution, and Heterologous Expression Nikolai Scherbak2, Anneli Ala-Häivälä2, Mikael Brosché2, Nathalie Böwer, Hilja Strid, John R. Gittins3, Elin Grahn, Leif A. Eriksson4, and Åke Strid* Akademin för naturvetenskap och teknik och Centrum för livsvetenskap (N.S., A.A.-H., N.B., E.G., L.A.E., Å.S) and Hälsoakademin och Centrum för livsvetenskap (H.S.), Örebro universitet, S-70182 Örebro, Sweden; Biokemi och biofysik (M.B., J.R.G.), Institutionen för Kemi, Göteborgs universitet, P.O. Box 462, S-405 30 Göteborg, Sweden; Division of Plant Biology (M.B.), Department of Biosciences, University of Helsinki, P.O. Box 65, FIN-00014 Helsinki, Finland, and Institute of Technology, University of Tartu, Nooruse 1, Tartu 50411, Estonia. 2 3 Footnotes: 1This work was supported by Helge Ax:son Johnson’s foundation (N.S.), the Carl Trygger Foundation (Å.S.) and the Faculty for Science and Technology at Örebro University (Å.S.). The Strategic Network for Swedish Plant Biotechnology (SSF) funded J.R.G.’s stay in Sweden with a grant to Å.S. -
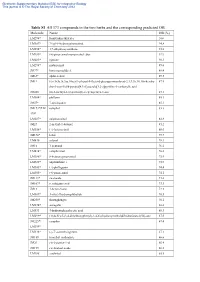
Table S1 All 373 Compounds in the Two Herbs and the Corresponding
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Table S1 All 373 compounds in the two herbs and the corresponding predicted OB Molecule Name OB (%) LM294* forsythidmethylester 100 LM387* 7'-epi-8-hydroxypinoresinol 94.8 LM394* 1,7-dihydroxyxanthone 93.8 LM289* (+)-pinoresinol monomethyl ether 92.9 LM285* egenine 90.3 LM298* matairesinol 89.6 JM77* loniceracetalide A 89.4 JM63* alpha-cedrol 89.3 JM1* (-)-(3r,8s,9r,9as,10as)-9-ethenyl-8-(beta-d-glucopyranosyloxy)-2,3,9,9a,10,10a-hexahy 87.5 dro-5-oxo-5h,8h-pyrano[4,3-d]oxazolo[3,2-a]pyridine-3-carboxylic acid JM200 (z)-3-methyl-2-(2-pentenyl)-2-cyclopenten-1-one 87.1 LM304* phillyrin 85.1 JM57* 7-epi-loganin 85.1 JM173*/LM camphol 83.3 398* LM287* epipinoresinol 82.8 JM27 2-methyl-1-butanol 81.2 LM356* (+)-lariciresinol 80.0 JM170* ledol 79.7 LM430 safynol 78.3 JM16 1-pentanol 76.2 LM414* campherenol 76.0 LM386* 8-hydroxypinoresinol 75.9 LM428* onjixanthone i 75.9 LM345* (+)-phillygenin 74.4 LM305* (+)-pinoresinol 74.1 JM113* sweroside 73.6 JM107* secologanic acid 73.3 JM13 1-hexen-3-one 72.3 LM309* 3-ethyl-7hydroxyphthalide 70.5 JM250* shuangkangsu 70.1 LM274* astragalin 68.6 LM331 4-hydroxyphenylacetic acid 68.3 LM299* (3r,4s,5r)-5-(3,4-dimethoxyphenyl)-3,4-bis(hydroxymethyl)dihydrofuran-2(3h)-one 67.5 JM225*/ camphor 67.4 LM399* LM338* (-)-7’-o-methylegenine 67.1 JM189 trimethyl isothiazole 66.6 JM29 cis-2-penten-1-ol 66.4 JM199 cis-linalool oxide 66.2 LM288 erythritol 65.8 Electronic Supplementary Material (ESI) for -

NPC Natural Product Communications
NPC-CMAPSEEC Issue Volume 12. Issue 2. Pages 153-318. 2017 ISSN 1934-578X (printed); ISSN 1555-9475 (online) www.naturalproduct.us NPC Natural Product Communications EDITOR-IN-CHIEF HONORARY EDITOR DR. PAWAN K AGRAWAL PROFESSOR GERALD BLUNDEN Natural Product Inc. The School of Pharmacy & Biomedical Sciences, 7963, Anderson Park Lane, University of Portsmouth, Westerville, Ohio 43081, USA Portsmouth, PO1 2DT U.K. [email protected] [email protected] EDITORS ADVISORY BOARD PROFESSOR ALEJANDRO F. BARRERO Department of Organic Chemistry, University of Granada, Prof. Giovanni Appendino Prof. Niel A. Koorbanally Campus de Fuente Nueva, s/n, 18071, Granada, Spain Novara, Italy Durban, South Africa [email protected] Prof. Norbert Arnold Prof. Chiaki Kuroda PROFESSOR MAURIZIO BRUNO Halle, Germany Tokyo, Japan Department STEBICEF, Prof. Hartmut Laatsch University of Palermo, Viale delle Scienze, Prof. Yoshinori Asakawa Gottingen, Germany Parco d’Orleans II - 90128 Palermo, Italy Tokushima, Japan [email protected] Prof. Marie Lacaille-Dubois Prof. Vassaya Bankova PROFESSOR VLADIMIR I. KALININ Dijon, France Sofia, Bulgaria G.B. Elyakov Pacific Institute of Bioorganic Chemistry, Prof. Shoei-Sheng Lee Far Eastern Branch, Russian Academy of Sciences, Prof. Roberto G. S. Berlinck Taipei, Taiwan Pr. 100-letya Vladivostoka 159, 690022, São Carlos, Brazil Vladivostok, Russian Federation Prof. Anna R. Bilia Prof. M. Soledade C. Pedras [email protected] Saskatoon, Canada Florence, Italy PROFESSOR YOSHIHIRO MIMAKI Prof. Geoffrey Cordell Prof. Luc Pieters School of Pharmacy, Antwerp, Belgium Chicago, IL, USA Tokyo University of Pharmacy and Life Sciences, Prof. Fatih Demirci Prof. Peter Proksch Horinouchi 1432-1, Hachioji, Tokyo 192-0392, Japan Düsseldorf, Germany [email protected] Eskişehir, Turkey Prof. -

Monoterpene Biosynthesis in the Glandulartrichomes Of
Plant Physiol. (1989) 89, 1351-1357 Received for publication July 1, 1988 0032-0889/0000/1351 /07/$01 .00/0 and in revised form December 2, 1988 Biochemical and Histochemical Localization of Monoterpene Biosynthesis in the Glandular Trichomes of Spearmint (Mentha spicata)" 2 Jonathan Gershenzon, Massimo Maffei3, and Rodney Croteau* Institute of Biological Chemistry, Washington State University, Pullman, Washington 99164-6340 ABSTRACT type diterpenes found in the glandular exudate. The head cells The primary monoterpene accumulated in the glandular tri- oftobacco trichomes are the only leafcells able to incorporate chomes of spearmint (Mentha spicata) is the ketone (-)-carvone radiolabeled acetate into duvane diterpenes; removal ofthese which is formed by cyclization of the C10 isoprenoid intermediate cells severely reduces or eliminates duvane production (22). geranyl pyrophosphate to the olefin (-)-limonene, hydroxylation Thus, it is tempting to speculate that all terpenes found in to (-)-trans-carveol and subsequent dehydrogenation. Selective glandular trichomes are synthesized in situ. However, recent extraction of the contents of the glandular trichomes indicated reports of monoterpene synthesis by undifferentiated cells in that essentially all of the cyclase and hydroxylase activities culture (5, 28), coupled to evidence for monoterpene transport resided in these structures, whereas only about 30% of the in intact plants (7, 31), suggest that the site of synthesis may carveol dehydrogenase was located here with the remainder not necessarily be the same as the site of accumulation. located in the rest of the leaf. This distribution of carveol dehy- drogenase activity was confirmed by histochemical methods. Spearmint (Mentha spicata, Lamiaceae) accumulates large Electrophoretic analysis of the partially purified carveol dehydro- quantities of monoterpenes in glandular trichomes, the major genase from extracts of both the glands and the leaves following constituent of which is (-)-carvone (20). -

Transcriptome Profiling of the Rice Blast Fungus During Invasive Plant
Mathioni et al. BMC Genomics 2011, 12:49 http://www.biomedcentral.com/1471-2164/12/49 RESEARCHARTICLE Open Access Transcriptome profiling of the rice blast fungus during invasive plant infection and in vitro stresses Sandra M Mathioni1, André Beló1, Christopher J Rizzo2, Ralph A Dean3, Nicole M Donofrio1* Abstract Background: Rice blast is the most threatening disease to cultivated rice. Magnaporthe oryzae, its causal agent, is likely to encounter environmental challenges during invasive growth in its host plants that require shifts in gene expression to establish a compatible interaction. Here, we tested the hypothesis that gene expression patterns during in planta invasive growth are similar to in vitro stress conditions, such as nutrient limitation, temperature up shift and oxidative stress, and determined which condition most closely mimicked that of in planta invasive growth. Gene expression data were collected from these in vitro experiments and compared to fungal gene expression during the invasive growth phase at 72 hours post-inoculation in compatible interactions on two grass hosts, rice and barley. Results: We identified 4,973 genes that were differentially expressed in at least one of the in planta and in vitro stress conditions when compared to fungal mycelia grown in complete medium, which was used as reference. From those genes, 1,909 showed similar expression patterns between at least one of the in vitro stresses and rice and/or barley. Hierarchical clustering of these 1,909 genes showed three major clusters in which in planta conditions closely grouped with the nutrient starvation conditions. Out of these 1,909 genes, 55 genes and 129 genes were induced and repressed in all treatments, respectively. -

Osmotic Stress Response in the Industrially Important Bacterium
Osmotic stress response in the industrially important bacterium Gluconobacter oxydans Dissertation to obtain the degree of Doctorate (Dr. rer. nat.) from the Faculty of Mathematics and Natural Sciences of the Rheinische Friedrich-Wilhelms University of Bonn, Germany submitted by Nageena Zahid from Lahore, Pakistan Bonn, November 2016 Finalized with the permission from the Faculty of Mathematics and Natural Sciences of the Rheinische Friedrich-Wilhelms University of Bonn, Germany First Referee: Prof. Dr. Uwe Deppenmeier Second Referee: Prof. Dr. Erwin A. Galinski Day of Promotion: 07.02.2017 Year of Publication: 2017 List of parts of this thesis that have already been published. Zahid N., Schweiger P., Galinski E., Deppenmeier U. (2015). Identification of mannitol as compatible solute in Gluconobacter oxydans. Appl Microbiol Biotechnol 99, 5511- 5521. Zahid N. and Deppenmeier U. (2016). Role of mannitol dehydrogenases in osmoprotection of Gluconobacter oxydans. Appl Microbiol Biotechnol 100, 9967-9978. For my Son TABLE OF CONTENTS 1. INTRODUCTION 1 1.1. Acetic acid bacteria 1 1.2. The genus Gluconobacter 2 1.2.1. Respiratory chain of G. oxydans 3 1.2.2. Intracellular carbohydrate metabolism in G. oxydans 6 1.2.3. Types and function of sugars and polyols metabolizing enzymes in Gluconobacter 7 1.2.4. Biotechnological applications of G. oxydans and its limitations 8 1.3. Aims of the work 10 2. MATERIALS AND METHODS 12 2.1. Chemicals and Enzymes 12 2.2. Bacterial strains, plasmids, primers 12 2.2.1. Bacterial strains 12 2.2.2. Oligonucleotides and Plasmids 13 2.3. Antibiotic stock solutions 18 2.4. -

Frankincense Essential Oil Prepared from Hydrodistillation of Boswellia
Ni et al. BMC Complementary and Alternative Medicine 2012, 12:253 http://www.biomedcentral.com/1472-6882/12/253 RESEARCH ARTICLE Open Access Frankincense essential oil prepared from hydrodistillation of Boswellia sacra gum resins induces human pancreatic cancer cell death in cultures and in a xenograft murine model Xiao Ni1, Mahmoud M Suhail2, Qing Yang3, Amy Cao4, Kar-Ming Fung3,5,6, Russell G Postier7, Cole Woolley8, Gary Young8, Jingzhe Zhang1* and Hsueh-Kung Lin3* Abstract Background: Regardless of the availability of therapeutic options, the overall 5-year survival for patients diagnosed with pancreatic cancer remains less than 5%. Gum resins from Boswellia species, also known as frankincense, have been used as a major ingredient in Ayurvedic and Chinese medicine to treat a variety of health-related conditions. Both frankincense chemical extracts and essential oil prepared from Boswellia species gum resins exhibit anti-neoplastic activity, and have been investigated as potential anti-cancer agents. The goals of this study are to identify optimal condition for preparing frankincense essential oil that possesses potent anti-tumor activity, and to evaluate the activity in both cultured human pancreatic cancer cells and a xenograft mouse cancer model. Methods: Boswellia sacra gum resins were hydrodistilled at 78°C; and essential oil distillate fractions were collected at different durations (Fraction I at 0–2 h, Fraction II at 8–10 h, and Fraction III at 11–12 h). Hydrodistillation of the second half of gum resins was performed at 100°C; and distillate was collected at 11–12 h (Fraction IV). Chemical compositions were identified by gas chromatography–mass spectrometry (GC-MS); and total boswellic acids contents were quantified by high-performance liquid chromatography (HPLC).