MVN: an R Package for Assessing Multivariate Normality
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Section 13 Kolmogorov-Smirnov Test
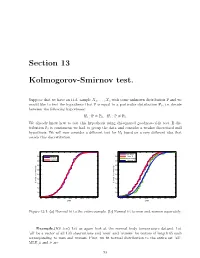
Section 13 Kolmogorov-Smirnov test. Suppose that we have an i.i.d. sample X1; : : : ; Xn with some unknown distribution P and we would like to test the hypothesis that P is equal to a particular distribution P0; i.e. decide between the following hypotheses: H : P = P ; H : P = P : 0 0 1 6 0 We already know how to test this hypothesis using chi-squared goodness-of-fit test. If dis tribution P0 is continuous we had to group the data and consider a weaker discretized null hypothesis. We will now consider a different test for H0 based on a very different idea that avoids this discretization. 1 1 men data 0.9 0.9 'men' fit all data women data 0.8 normal fit 0.8 'women' fit 0.7 0.7 0.6 0.6 0.5 0.5 0.4 0.4 Cumulative probability 0.3 Cumulative probability 0.3 0.2 0.2 0.1 0.1 0 0 96 96.5 97 97.5 98 98.5 99 99.5 100 100.5 96.5 97 97.5 98 98.5 99 99.5 100 100.5 Data Data Figure 13.1: (a) Normal fit to the entire sample. (b) Normal fit to men and women separately. Example.(KS test) Let us again look at the normal body temperature dataset. Let ’all’ be a vector of all 130 observations and ’men’ and ’women’ be vectors of length 65 each corresponding to men and women. First, we fit normal distribution to the entire set ’all’. -
Linear Regression in SPSS
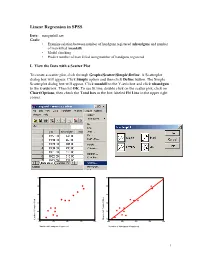
Linear Regression in SPSS Data: mangunkill.sav Goals: • Examine relation between number of handguns registered (nhandgun) and number of man killed (mankill) • Model checking • Predict number of man killed using number of handguns registered I. View the Data with a Scatter Plot To create a scatter plot, click through Graphs\Scatter\Simple\Define. A Scatterplot dialog box will appear. Click Simple option and then click Define button. The Simple Scatterplot dialog box will appear. Click mankill to the Y-axis box and click nhandgun to the x-axis box. Then hit OK. To see fit line, double click on the scatter plot, click on Chart\Options, then check the Total box in the box labeled Fit Line in the upper right corner. 60 60 50 50 40 40 30 30 20 20 10 10 Killed of People Number Number of People Killed 400 500 600 700 800 400 500 600 700 800 Number of Handguns Registered Number of Handguns Registered 1 Click the target button on the left end of the tool bar, the mouse pointer will change shape. Move the target pointer to the data point and click the left mouse button, the case number of the data point will appear on the chart. This will help you to identify the data point in your data sheet. II. Regression Analysis To perform the regression, click on Analyze\Regression\Linear. Place nhandgun in the Dependent box and place mankill in the Independent box. To obtain the 95% confidence interval for the slope, click on the Statistics button at the bottom and then put a check in the box for Confidence Intervals. -

A Robust Rescaled Moment Test for Normality in Regression
Journal of Mathematics and Statistics 5 (1): 54-62, 2009 ISSN 1549-3644 © 2009 Science Publications A Robust Rescaled Moment Test for Normality in Regression 1Md.Sohel Rana, 1Habshah Midi and 2A.H.M. Rahmatullah Imon 1Laboratory of Applied and Computational Statistics, Institute for Mathematical Research, University Putra Malaysia, 43400 Serdang, Selangor, Malaysia 2Department of Mathematical Sciences, Ball State University, Muncie, IN 47306, USA Abstract: Problem statement: Most of the statistical procedures heavily depend on normality assumption of observations. In regression, we assumed that the random disturbances were normally distributed. Since the disturbances were unobserved, normality tests were done on regression residuals. But it is now evident that normality tests on residuals suffer from superimposed normality and often possess very poor power. Approach: This study showed that normality tests suffer huge set back in the presence of outliers. We proposed a new robust omnibus test based on rescaled moments and coefficients of skewness and kurtosis of residuals that we call robust rescaled moment test. Results: Numerical examples and Monte Carlo simulations showed that this proposed test performs better than the existing tests for normality in the presence of outliers. Conclusion/Recommendation: We recommend using our proposed omnibus test instead of the existing tests for checking the normality of the regression residuals. Key words: Regression residuals, outlier, rescaled moments, skewness, kurtosis, jarque-bera test, robust rescaled moment test INTRODUCTION out that the powers of t and F tests are extremely sensitive to the hypothesized error distribution and may In regression analysis, it is a common practice over deteriorate very rapidly as the error distribution the years to use the Ordinary Least Squares (OLS) becomes long-tailed. -

Downloaded 09/24/21 09:09 AM UTC
MARCH 2007 NOTES AND CORRESPONDENCE 1151 NOTES AND CORRESPONDENCE A Cautionary Note on the Use of the Kolmogorov–Smirnov Test for Normality DAG J. STEINSKOG Nansen Environmental and Remote Sensing Center, and Bjerknes Centre for Climate Research, and Geophysical Institute, University of Bergen, Bergen, Norway DAG B. TJØSTHEIM Department of Mathematics, University of Bergen, Bergen, Norway NILS G. KVAMSTØ Geophysical Institute, University of Bergen, and Bjerknes Centre for Climate Research, Bergen, Norway (Manuscript received and in final form 28 April 2006) ABSTRACT The Kolmogorov–Smirnov goodness-of-fit test is used in many applications for testing normality in climate research. This note shows that the test usually leads to systematic and drastic errors. When the mean and the standard deviation are estimated, it is much too conservative in the sense that its p values are strongly biased upward. One may think that this is a small sample problem, but it is not. There is a correction of the Kolmogorov–Smirnov test by Lilliefors, which is in fact sometimes confused with the original Kolmogorov–Smirnov test. Both the Jarque–Bera and the Shapiro–Wilk tests for normality are good alternatives to the Kolmogorov–Smirnov test. A power comparison of eight different tests has been undertaken, favoring the Jarque–Bera and the Shapiro–Wilk tests. The Jarque–Bera and the Kolmogorov– Smirnov tests are also applied to a monthly mean dataset of geopotential height at 500 hPa. The two tests give very different results and illustrate the danger of using the Kolmogorov–Smirnov test. 1. Introduction estimates of skewness and kurtosis. Such moments can be used to diagnose nonlinear processes and to provide The Kolmogorov–Smirnov test (hereafter the KS powerful tools for validating models (see also Ger- test) is a much used goodness-of-fit test. -

Testing Normality: a GMM Approach Christian Bontempsa and Nour
Testing Normality: A GMM Approach Christian Bontempsa and Nour Meddahib∗ a LEEA-CENA, 7 avenue Edouard Belin, 31055 Toulouse Cedex, France. b D´epartement de sciences ´economiques, CIRANO, CIREQ, Universit´ede Montr´eal, C.P. 6128, succursale Centre-ville, Montr´eal (Qu´ebec), H3C 3J7, Canada. First version: March 2001 This version: June 2003 Abstract In this paper, we consider testing marginal normal distributional assumptions. More precisely, we propose tests based on moment conditions implied by normality. These moment conditions are known as the Stein (1972) equations. They coincide with the first class of moment conditions derived by Hansen and Scheinkman (1995) when the random variable of interest is a scalar diffusion. Among other examples, Stein equation implies that the mean of Hermite polynomials is zero. The GMM approach we adopt is well suited for two reasons. It allows us to study in detail the parameter uncertainty problem, i.e., when the tests depend on unknown parameters that have to be estimated. In particular, we characterize the moment conditions that are robust against parameter uncertainty and show that Hermite polynomials are special examples. This is the main contribution of the paper. The second reason for using GMM is that our tests are also valid for time series. In this case, we adopt a Heteroskedastic-Autocorrelation-Consistent approach to estimate the weighting matrix when the dependence of the data is unspecified. We also make a theoretical comparison of our tests with Jarque and Bera (1980) and OPG regression tests of Davidson and MacKinnon (1993). Finite sample properties of our tests are derived through a comprehensive Monte Carlo study. -

Exponentiality Test Using a Modified Lilliefors Test Achut Prasad Adhikari
University of Northern Colorado Scholarship & Creative Works @ Digital UNC Dissertations Student Research 5-1-2014 Exponentiality Test Using a Modified Lilliefors Test Achut Prasad Adhikari Follow this and additional works at: http://digscholarship.unco.edu/dissertations Recommended Citation Adhikari, Achut Prasad, "Exponentiality Test Using a Modified Lilliefors Test" (2014). Dissertations. Paper 63. This Text is brought to you for free and open access by the Student Research at Scholarship & Creative Works @ Digital UNC. It has been accepted for inclusion in Dissertations by an authorized administrator of Scholarship & Creative Works @ Digital UNC. For more information, please contact [email protected]. UNIVERSITY OF NORTHERN COLORADO Greeley, Colorado The Graduate School AN EXPONENTIALITY TEST USING A MODIFIED LILLIEFORS TEST A Dissertation Submitted in Partial Fulfillment Of the Requirements for the Degree of Doctor of Philosophy Achut Prasad Adhikari College of Education and Behavioral Sciences Department of Applied Statistics and Research Methods May 2014 ABSTRACT Adhikari, Achut Prasad An Exponentiality Test Using a Modified Lilliefors Test. Published Doctor of Philosophy dissertation, University of Northern Colorado, 2014. A new exponentiality test was developed by modifying the Lilliefors test of exponentiality for the purpose of improving the power of the test it directly modified. Lilliefors has considered the maximum absolute differences between the sample empirical distribution function (EDF) and the exponential cumulative distribution function (CDF). The proposed test considered the sum of all the absolute differences between the CDF and EDF. By considering the sum of all the absolute differences rather than only a point difference of each observation, the proposed test would expect to be less affected by individual extreme (too low or too high) observations and capable of detecting smaller, but consistent, differences between the distributions. -

Power Comparisons of Shapiro-Wilk, Kolmogorov-Smirnov, Lilliefors and Anderson-Darling Tests
Journal ofStatistical Modeling and Analytics Vol.2 No.I, 21-33, 2011 Power comparisons of Shapiro-Wilk, Kolmogorov-Smirnov, Lilliefors and Anderson-Darling tests Nornadiah Mohd Razali1 Yap Bee Wah 1 1Faculty ofComputer and Mathematica/ Sciences, Universiti Teknologi MARA, 40450 Shah Alam, Selangor, Malaysia E-mail: nornadiah@tmsk. uitm. edu.my, yapbeewah@salam. uitm. edu.my ABSTRACT The importance of normal distribution is undeniable since it is an underlying assumption of many statistical procedures such as I-tests, linear regression analysis, discriminant analysis and Analysis of Variance (ANOVA). When the normality assumption is violated, interpretation and inferences may not be reliable or valid. The three common procedures in assessing whether a random sample of independent observations of size n come from a population with a normal distribution are: graphical methods (histograms, boxplots, Q-Q-plots), numerical methods (skewness and kurtosis indices) and formal normality tests. This paper* compares the power offour formal tests of normality: Shapiro-Wilk (SW) test, Kolmogorov-Smirnov (KS) test, Lillie/ors (LF) test and Anderson-Darling (AD) test. Power comparisons of these four tests were obtained via Monte Carlo simulation of sample data generated from alternative distributions that follow symmetric and asymmetric distributions. Ten thousand samples ofvarious sample size were generated from each of the given alternative symmetric and asymmetric distributions. The power of each test was then obtained by comparing the test of normality statistics with the respective critical values. Results show that Shapiro-Wilk test is the most powerful normality test, followed by Anderson-Darling test, Lillie/ors test and Kolmogorov-Smirnov test. However, the power ofall four tests is still low for small sample size. -

A Quantitative Validation Method of Kriging Metamodel for Injection Mechanism Based on Bayesian Statistical Inference
metals Article A Quantitative Validation Method of Kriging Metamodel for Injection Mechanism Based on Bayesian Statistical Inference Dongdong You 1,2,3,* , Xiaocheng Shen 1,3, Yanghui Zhu 1,3, Jianxin Deng 2,* and Fenglei Li 1,3 1 National Engineering Research Center of Near-Net-Shape Forming for Metallic Materials, South China University of Technology, Guangzhou 510640, China; [email protected] (X.S.); [email protected] (Y.Z.); fl[email protected] (F.L.) 2 Guangxi Key Lab of Manufacturing System and Advanced Manufacturing Technology, Guangxi University, Nanning 530003, China 3 Guangdong Key Laboratory for Advanced Metallic Materials processing, South China University of Technology, Guangzhou 510640, China * Correspondence: [email protected] (D.Y.); [email protected] (J.D.); Tel.: +86-20-8711-2933 (D.Y.); +86-137-0788-9023 (J.D.) Received: 2 April 2019; Accepted: 24 April 2019; Published: 27 April 2019 Abstract: A Bayesian framework-based approach is proposed for the quantitative validation and calibration of the kriging metamodel established by simulation and experimental training samples of the injection mechanism in squeeze casting. The temperature data uncertainty and non-normal distribution are considered in the approach. The normality of the sample data is tested by the Anderson–Darling method. The test results show that the original difference data require transformation for Bayesian testing due to the non-normal distribution. The Box–Cox method is employed for the non-normal transformation. The hypothesis test results of the calibrated kriging model are more reliable after data transformation. The reliability of the kriging metamodel is quantitatively assessed by the calculated Bayes factor and confidence. -

Swilk — Shapiro–Wilk and Shapiro–Francia Tests for Normality
Title stata.com swilk — Shapiro–Wilk and Shapiro–Francia tests for normality Description Quick start Menu Syntax Options for swilk Options for sfrancia Remarks and examples Stored results Methods and formulas Acknowledgment References Also see Description swilk performs the Shapiro–Wilk W test for normality for each variable in the specified varlist. Likewise, sfrancia performs the Shapiro–Francia W 0 test for normality. See[ MV] mvtest normality for multivariate tests of normality. Quick start Shapiro–Wilk test of normality Shapiro–Wilk test for v1 swilk v1 Separate tests of normality for v1 and v2 swilk v1 v2 Generate new variable w containing W test coefficients swilk v1, generate(w) Specify that average ranks should not be used for tied values swilk v1 v2, noties Test that v3 is distributed lognormally generate lnv3 = ln(v3) swilk lnv3 Shapiro–Francia test of normality Shapiro–Francia test for v1 sfrancia v1 Separate tests of normality for v1 and v2 sfrancia v1 v2 As above, but use the Box–Cox transformation sfrancia v1 v2, boxcox Specify that average ranks should not be used for tied values sfrancia v1 v2, noties 1 2 swilk — Shapiro–Wilk and Shapiro–Francia tests for normality Menu swilk Statistics > Summaries, tables, and tests > Distributional plots and tests > Shapiro-Wilk normality test sfrancia Statistics > Summaries, tables, and tests > Distributional plots and tests > Shapiro-Francia normality test Syntax Shapiro–Wilk normality test swilk varlist if in , swilk options Shapiro–Francia normality test sfrancia varlist if in , sfrancia options swilk options Description Main generate(newvar) create newvar containing W test coefficients lnnormal test for three-parameter lognormality noties do not use average ranks for tied values sfrancia options Description Main boxcox use the Box–Cox transformation for W 0; the default is to use the log transformation noties do not use average ranks for tied values by and collect are allowed with swilk and sfrancia; see [U] 11.1.10 Prefix commands. -
Univariate Analysis and Normality Test Using SAS, STATA, and SPSS
© 2002-2006 The Trustees of Indiana University Univariate Analysis and Normality Test: 1 Univariate Analysis and Normality Test Using SAS, STATA, and SPSS Hun Myoung Park This document summarizes graphical and numerical methods for univariate analysis and normality test, and illustrates how to test normality using SAS 9.1, STATA 9.2 SE, and SPSS 14.0. 1. Introduction 2. Graphical Methods 3. Numerical Methods 4. Testing Normality Using SAS 5. Testing Normality Using STATA 6. Testing Normality Using SPSS 7. Conclusion 1. Introduction Descriptive statistics provide important information about variables. Mean, median, and mode measure the central tendency of a variable. Measures of dispersion include variance, standard deviation, range, and interquantile range (IQR). Researchers may draw a histogram, a stem-and-leaf plot, or a box plot to see how a variable is distributed. Statistical methods are based on various underlying assumptions. One common assumption is that a random variable is normally distributed. In many statistical analyses, normality is often conveniently assumed without any empirical evidence or test. But normality is critical in many statistical methods. When this assumption is violated, interpretation and inference may not be reliable or valid. Figure 1. Comparing the Standard Normal and a Bimodal Probability Distributions Standard Normal Distribution Bimodal Distribution .4 .4 .3 .3 .2 .2 .1 .1 0 0 -5 -3 -1 1 3 5 -5 -3 -1 1 3 5 T-test and ANOVA (Analysis of Variance) compare group means, assuming variables follow normal probability distributions. Otherwise, these methods do not make much http://www.indiana.edu/~statmath © 2002-2006 The Trustees of Indiana University Univariate Analysis and Normality Test: 2 sense. -

2.4.3 Examining Skewness and Kurtosis for Normality Using SPSS We Will Analyze the Examples Earlier Using SPSS
24 CHAPTER 2 TEsTing DATA foR noRmAliTy Finally, use the skewness and the standard error of the skewness to ind a z-score: Sk −0− 1.018 zS = = k SE 0. 501 Sk 2. 032 zSk = − Use the z-score to examine the sample’s approximation to a normal distribution. This value must fall between −1.96 and +1.96 to pass the normality assumption for α = 0.05. Since this z-score value does not fall within that range, the sample has failed our normality assumption for skewness. Therefore, either the sample must be modiied and rechecked or you must use a nonparametric statistical test. 2.4.3 Examining Skewness and Kurtosis for Normality Using SPSS We will analyze the examples earlier using SPSS. 2.4.3.1 Deine Your Variables First, click the “Variable View” tab at the bottom of your screen. Then, type the name of your variable(s) in the “Name” column. As shown in Figure 2.7, we have named our variable “Wk1_Qz.” FIGURE 2.7 2.4.3.2 Type in Your Values Click the “Data View” tab at the bottom of your screen and type your data under the variable names. As shown in Figure 2.8, we have typed the values for the “Wk1_Qz” sample. 2.4.3.3 Analyze Your Data As shown in Figure 2.9, use the pull-down menus to choose “Analyze,” “Descriptive Statistics,” and “Descriptives . .” Choose the variable(s) that you want to examine. Then, click the button in the middle to move the variable to the “Variable(s)” box, as shown in Figure 2.10. -

Normality Tests (Simulation)
PASS Sample Size Software NCSS.com Chapter 670 Normality Tests (Simulation) Introduction This procedure allows you to study the power and sample size of eight statistical tests of normality. Since there are no formulas that allow the calculation of power directly, simulation is used. This gives you the ability to compare the adequacy of each test under a wide variety of solutions. The reason there are so many different normality tests is that there are many different forms of normality. Thode (2002) presents that following recommendations concerning which tests to use for each situation. Note that the details of each test will be presented later. Normal vs. Long-Tailed Symmetric Alternative Distributions The Shapiro-Wilk and the kurtosis tests have been found to be best for normality testing against long-tailed symmetric alternatives. Normal vs. Short-Tailed Symmetric Alternative Distributions The Shapiro-Wilk and the range tests have been found to be best for normality testing against short-tailed symmetric alternatives. Normal vs. Asymmetric Alternative Distributions The Shapiro-Wilk and the skewness tests have been found to be best for normality testing against asymmetric alternatives. Technical Details Computer simulation allows one to estimate the power and significance level that is actually achieved by a test procedure in situations that are not mathematically tractable. Computer simulation was once limited to mainframe computers. Currently, due to increased computer speeds, simulation studies can be completed on desktop and laptop computers in a reasonable period of time. The steps to a simulation study are as follows. 1. Specify which the normality test is to be used.