Effects of Simulated Microgravity on the Proteome and Secretome of the Polyextremotolerant Black Fungus Knufia Chersonesos
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evaluating Feruloyl Esterase—Xylanase Synergism For
agronomy Article Evaluating Feruloyl Esterase—Xylanase Synergism for Hydroxycinnamic Acid and Xylo-Oligosaccharide Production from Untreated, Hydrothermally Pre-Treated and Dilute-Acid Pre-Treated Corn Cobs Lithalethu Mkabayi 1 , Samkelo Malgas 1 , Brendan S. Wilhelmi 2 and Brett I. Pletschke 1,* 1 Enzyme Science Programme (ESP), Department of Biochemistry and Microbiology, Rhodes University, Grahamstown, Eastern Cape 6140, South Africa; [email protected] (L.M.); [email protected] (S.M.) 2 Department of Biochemistry and Microbiology, Rhodes University, Grahamstown, Eastern Cape 6140, South Africa; [email protected] * Correspondence: [email protected]; Tel.: +27-46-6038081 Received: 4 April 2020; Accepted: 30 April 2020; Published: 13 May 2020 Abstract: Agricultural residues are considered the most promising option as a renewable feedstock for biofuel and high valued-added chemical production due to their availability and low cost. The efficient enzymatic hydrolysis of agricultural residues into value-added products such as sugars and hydroxycinnamic acids is a challenge because of the recalcitrant properties of the native biomass. Development of synergistic enzyme cocktails is required to overcome biomass residue recalcitrance, and achieve high yields of potential value-added products. In this study, the synergistic action of two termite metagenome-derived feruloyl esterases (FAE5 and FAE6), and an endo-xylanase (Xyn11) from Thermomyces lanuginosus, was optimized using 0.5% (w/v) insoluble wheat arabinoxylan (a model substrate) and then applied to 1% (w/v) corn cobs for the efficient production of xylo-oligosaccharides (XOS) and hydroxycinnamic acids. The enzyme combination of 66% Xyn11 and 33% FAE5 or FAE6 (protein loading) produced the highest amounts of XOS, ferulic acid, and p-coumaric acid from untreated, hydrothermal, and acid pre-treated corn cobs. -

43-09-405 and 407 Final Combine
Pak. J. Bot ., 44(6): 2093-2102, 2012. A NEW SPECIES OF TIAROSPORELLA AZADARICHTA AND NEW FUNGAL RECORDS ON AZADIRACHTA INDICA FROM PAKISTAN SYED QAISER ABBAS 1, TEHREEMA IFTIKHAR 1* , MUBASHIR NIAZ 1, IFTIKHAR ALI 1, NABILA IFTIKHAR 1 AND ALIA ABBAS 2 1Department of Botany, Government College University, Alama Iqbal Road, Faisalabad, Pakistan 2Department of Botany, Federal Urdu University, Karachi, Pakistan *Corresponding author’s e-mail: [email protected] Abstract A new species of Tiarosporella azadarichta has been described on Azadirachta indica and some new fungal records viz ., Diplozythiella bambusina , Ulocladium chartarum , Cladosporium nigrellum, Cladosporium oxysporum. Didymostilbe coffeae , Muellerella pygmaea, Lasiodiplodia paraphysaria, Monochaetinula terminalae, Trimmatostroma sp., and Epidermophyton floccosum are reported on Azadirachta indica for the first time from Pakistan. Introduction reason a detailed survey of the plant for fungi was under taken in this study. Azadirachta indica (as Azadirachta Juss, Melia azadirachta ) belong to the family Meliaceae. Neem is its Materials and Methods common name. It is important due to its commercial and medicinal value. Azadirachta indica is also an important Samples of Azadirachta indica were collected from plant for its potential of anti-fungal, anti-bacterial and the different areas of District Faisalabad and Gojra. The insecticidal activity. Decline of trees due to fungi are different areas were G.C. University. Faisalabad, increasing tremendously in Pakistan especially in Punjab Agriculture University Faisalabad, Gutwala forest (park) and Sindh (Javed et al ., 2004; Khanzada et al ., 2011; Fateh Faisalabad, Tandlianwala and Gojra City. Methods and et al., 2011; Rheman et al., 2011; Farooq et al., 2011). materials are the same as described Abbas et al ., (2010). -

Kramers' Theory and the Dependence of Enzyme Dynamics on Trehalose
catalysts Review Kramers’ Theory and the Dependence of Enzyme Dynamics on Trehalose-Mediated Viscosity José G. Sampedro 1,* , Miguel A. Rivera-Moran 1 and Salvador Uribe-Carvajal 2 1 Instituto de Física, Universidad Autónoma de San Luis Potosí, Manuel Nava 6, Zona Universitaria, San Luis Potosí C.P. 78290, Mexico; [email protected] 2 Instituto de Fisiología Celular, Universidad Nacional Autónoma de México, Ciudad de México C.P. 04510, Mexico; [email protected] * Correspondence: [email protected]fisica.uaslp.mx; Tel.: +52-(444)-8262-3200 (ext. 5715) Received: 29 April 2020; Accepted: 29 May 2020; Published: 11 June 2020 Abstract: The disaccharide trehalose is accumulated in the cytoplasm of some organisms in response to harsh environmental conditions. Trehalose biosynthesis and accumulation are important for the survival of such organisms by protecting the structure and function of proteins and membranes. Trehalose affects the dynamics of proteins and water molecules in the bulk and the protein hydration shell. Enzyme catalysis and other processes dependent on protein dynamics are affected by the viscosity generated by trehalose, as described by the Kramers’ theory of rate reactions. Enzyme/protein stabilization by trehalose against thermal inactivation/unfolding is also explained by the viscosity mediated hindering of the thermally generated structural dynamics, as described by Kramers’ theory. The analysis of the relationship of viscosity–protein dynamics, and its effects on enzyme/protein function and other processes (thermal inactivation and unfolding/folding), is the focus of the present work regarding the disaccharide trehalose as the viscosity generating solute. Finally, trehalose is widely used (alone or in combination with other compounds) in the stabilization of enzymes in the laboratory and in biotechnological applications; hence, considering the effect of viscosity on catalysis and stability of enzymes may help to improve the results of trehalose in its diverse uses/applications. -

An Unusual Haemoid Fungi: Ulocladium, As a Cause Of
Volume 2 Number 2 (June 2010) 95-97 CASE Report An unusual phaeoid fungi: Ulocladium, as a cause of chronic allergic fungal sinusitis Kaur R, Wadhwa A1,Gulati A2, Agrawal AK2 1Department of Microbiology. 2Department of Otorhinolaryngology, Maulana Azad Medical College, New Delhi, India. Received: April 2010, Accepted: May 2010. ABSTRACT Allergic fungal sinusitis (AFS) has been recognized as an important cause of chronic sinusitis commonly caused by Aspergillus spp. and various dematiaceous fungi like Bipolaris, Alternaria, Curvalaria, and etc. Ulocladium botrytis is a non pathogenic environmental dematiaceous fungi, which has been recently described as a human pathogen. Ulocladium has never been associated with allergic fungal sinusitis but it was identified as an etiological agent of AFS in a 35 year old immunocompetent female patient presenting with chronic nasal obstruction of several months duration to our hospital. The patient underwent FESS and the excised polyps revealed Ulocladium as the causative fungal agent. Keywords: Ulocladium, Phaeoid Fungi, Chronic Sinusitis. INTRODUCTION CASE REPORT Chronic allergic sinusitis is a common condition A 35 year old female patient presented to the ENT responsible for the development of nasal polyps, out patient department of the Lok Nayak Hospital, described as abnormal lesions that emanate from Delhi, with chronic nasal obstruction, excessive any portion of the nasal mucosa or Para nasal sneezing, nasal discharge and frontal headache since sinuses. They are commonly located in the middle several months. Nasal obstruction was of gradual meatus and ethmoid sinus and are present in onset, non progressive, more on the left side than 1-4% of the population (1). -

Feruloyl Esterases: Biocatalysts to Overcome Biomass Recalcitrance and for the Production of Bioactive Compounds Dyoni M
Feruloyl esterases: Biocatalysts to overcome biomass recalcitrance and for the production of bioactive compounds Dyoni M. Oliveira, Thatiane R. Mota, Bianca Oliva, Fernando Segato, Rogério Marchiosi, Osvaldo Ferrarese-Filho, Craig Faulds, Wanderley D. dos Santos To cite this version: Dyoni M. Oliveira, Thatiane R. Mota, Bianca Oliva, Fernando Segato, Rogério Marchiosi, et al.. Feru- loyl esterases: Biocatalysts to overcome biomass recalcitrance and for the production of bioactive com- pounds. Bioresource Technology, Elsevier, 2019, 278, pp.408-423. 10.1016/j.biortech.2019.01.064. hal-02627378 HAL Id: hal-02627378 https://hal.inrae.fr/hal-02627378 Submitted on 26 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Accepted Manuscript Review Feruloyl esterases: Biocatalysts to overcome biomass recalcitrance and for the production of bioactive compounds Dyoni M. Oliveira, Thatiane R. Mota, Bianca Oliva, Fernando Segato, Rogério Marchiosi, Osvaldo Ferrarese-Filho, Craig -
Cutaneous Infection Caused by Ulocladium Chartarum in a Heart Transplant Recipient: Case Report and Review
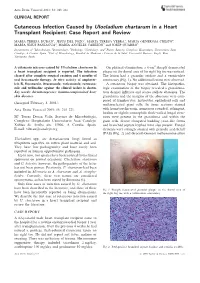
Acta Derm Venereol 2003; 83: 218–221 CLINICAL REPORT Cutaneous Infection Caused by Ulocladium chartarum in a Heart Transplant Recipient: Case Report and Review MARI´A TERESA DURA´ N1, JESU´ S DEL POZO2, MARI´A TERESA YEBRA3, MARI´A GENEROSA CRESPO4, MARI´A JESU´ S PANIAGUA4, MARI´A ANGELES CABEZO´ N5 and JOSEP GUARRO6 Departments of 1Microbiology, 2Dermatology, 3Pathology, 4Cardiology, and 5Plastic Surgery, Complexo Hospitalario Universitario Juan Canalejo, A Corun˜a, Spain, 6Unit of Microbiology, Facultat de Medicina i Cie`ncies de la Salut, Universitat Rovira i Virgili, Reus, Tarragona, Spain A cutaneous mycoses caused by Ulocladium chartarum in On physical examination, a 6-cm2 sharply demarcated a heart transplant recipient is reported. The infection plaque on the dorsal area of his right big toe was noticed. cleared after complete surgical excision and 6 months of The lesion had a granular surface and a vermiculate oral itraconazole therapy. In vitro activity of amphoter- consistency (Fig. 1). No additional lesions were observed. icin B, fluconazole, itraconazole, voriconazole, ravucona- A cutaneous biopsy was obtained. The histopatho- zole and terbinafine against the clinical isolate is shown. logic examination of the biopsy revealed a granuloma- Key words: dermatomycoses; immunocompromised host; tous dermal infiltrate and scarce stellate abscesses. The skin diseases. granuloma and the margins of the abscesses were com- posed of lymphocytes, histiocytes, epithelioid cells and (Accepted February 3, 2003.) multinucleated giant cells. In tissue sections stained Acta Derm Venereol 2003; 83: 218–221. with hematoxylin-eosin, numerous rounded, refringent, hyaline or slightly eosinophilic thick-walled fungal struc- Ma Teresa Dura´n Valle, Servicio de Microbiologı´a, tures were present in the granuloma and within the Complexo Hospitalario Universitario Juan Canalejo, giant cells. -

Cutinase Structure, Function and Biocatalytic Applications
EJB Electronic Journal of Biotechnology ISSN: 0717-345 Vol.1 No.3, Issue of August 15, 1998. © 1998 by Universidad Católica de Valparaíso –- Chile Invited review paper/ Received 20 October, 1998. REVIEW ARTICLE Cutinase structure, function and biocatalytic applications Cristina M. L. Carvalho Centro de Engenharia Biológica e Química Instituto Superior Técnico 1000 Lisboa – Portugal Maria Raquel Aires-Barros Centro de Engenharia Biológica e Química Instituto Superior Técnico 1000 Lisboa – Portugal 1 Joaquim M. S. Cabral Centro de Engenharia Biológica e Química Instituto Superior Técnico 1000 Lisboa – Portugal Tel: + 351.1.8419065 Fax: + 351.1.8419062 E-mail: [email protected] http://www.ist.utl.pt/ This review analyses the role of cutinases in nature and Some microorganisms, including plant pathogens, have their potential biotechnological applications. The been shown to live on cutin as their sole carbon source cloning and expression of a fungal cutinase from (Purdy and Kolattukudy, 1975) and the production of Fusarium solani f. pisi, in Escherichia coli and extracellular cutinolytic enzymes has been presented (Purdy Saccharomyces cerevisiae hosts are described. The three and Kolattukudy, 1975; Lin and Kolattukudy, 1980). dimensional structure of this cutinase is also analysed Cutinases have been purified and characterized from and its function as a lipase discussed and compared with several different sources, mainly fungi (Purdy and other lipases. The biocatalytic applications of cutinase Kolattukudy, 1975 ; Lin and Kolattukudy, 1980; Petersen et are described taking into account the preparation of al., 1997; Dantzig et al., 1986; Murphy et al., 1996) and different cutinase forms and the media where the pollen (Petersen et al., 1997; Sebastian et al., 1987). -

Culture-Dependent and Amplicon Sequencing Approaches Reveal Diversity and Distribution of Black Fungi in Antarctic Cryptoendolithic Communities
Journal of Fungi Article Culture-Dependent and Amplicon Sequencing Approaches Reveal Diversity and Distribution of Black Fungi in Antarctic Cryptoendolithic Communities Laura Selbmann 1,2, Gerardo A. Stoppiello 1, Silvano Onofri 1 , Jason E. Stajich 3,* and Claudia Coleine 1,* 1 Department of Ecological and Biological Sciences, University of Tuscia, 01100 Viterbo, Italy; [email protected] (L.S.); [email protected] (G.A.S.); [email protected] (S.O.) 2 Mycological Section, Italian Antarctic National Museum (MNA), 16121 Genoa, Italy 3 Department of Microbiology and Plant Pathology, University of California, Riverside, CA 92521, USA * Correspondence: [email protected] (J.E.S.); [email protected] (C.C.); Tel.: +1-951-827-2363 (J.E.S.); +39-0761-357138 (C.C.) Abstract: In the harshest environmental conditions of the Antarctic desert, normally incompatible with active life, microbes are adapted to exploit the cryptoendolithic habitat (i.e., pore spaces of rocks) and represent the predominant life-forms. In the rocky niche, microbes take advantage of the thermal buffering, physical stability, protection against UV radiation, excessive solar radiation, and water retention—of paramount importance in one of the driest environments on Earth. In this work, high-throughput sequencing and culture-dependent approaches have been combined, for the first time, to untangle the diversity and distribution of black fungi in the Antarctic cryptoendolithic microbial communities, hosting some of the most extreme-tolerant microorganisms. Rock samples were collected in a vast area, along an altitudinal gradient and opposite sun exposure—known to influence microbial diversity—with the aim to compare and integrate results gained with the two Citation: Selbmann, L.; Stoppiello, Friedmanniomyces endolithicus G.A.; Onofri, S.; Stajich, J.E.; Coleine, approaches. -

Cutinases from Mycobacterium Tuberculosis
Identification of Residues Involved in Substrate Specificity and Cytotoxicity of Two Closely Related Cutinases from Mycobacterium tuberculosis Luc Dedieu, Carole Serveau-Avesque, Ste´phane Canaan* CNRS - Aix-Marseille Universite´ - Enzymologie Interfaciale et Physiologie de la Lipolyse - UMR 7282, Marseille, France Abstract The enzymes belonging to the cutinase family are serine enzymes active on a large panel of substrates such as cutin, triacylglycerols, and phospholipids. In the M. tuberculosis H37Rv genome, seven genes coding for cutinase-like proteins have been identified with strong immunogenic properties suggesting a potential role as vaccine candidates. Two of these enzymes which are secreted and highly homologous, possess distinct substrates specificities. Cfp21 is a lipase and Cut4 is a phospholipase A2, which has cytotoxic effects on macrophages. Structural overlay of their three-dimensional models allowed us to identify three areas involved in the substrate binding process and to shed light on this substrate specificity. By site-directed mutagenesis, residues present in these Cfp21 areas were replaced by residues occurring in Cut4 at the same location. Three mutants acquired phospholipase A1 and A2 activities and the lipase activities of two mutants were 3 and 15 fold greater than the Cfp21 wild type enzyme. In addition, contrary to mutants with enhanced lipase activity, mutants that acquired phospholipase B activities induced macrophage lysis as efficiently as Cut4 which emphasizes the relationship between apparent phospholipase A2 activity and cytotoxicity. Modification of areas involved in substrate specificity, generate recombinant enzymes with higher activity, which may be more immunogenic than the wild type enzymes and could therefore constitute promising candidates for antituberculous vaccine production. -

Purification and Characterization of Cutinase from Venturia Inaequalis
Physiology and Biochemistry Purification and Characterization of Cutinase from Venturia inaequalis Wolfram K611er and Diana M. Parker Assistant professor and research assistant, Department of Plant Pathology, Cornell University, New York State Agricultural Experiment Station, Geneva 14456. We thank Professor A. L. Jones for the isolates of Venturia inaequalis, Mr. Robert W. Ennis, Jr. for his help in cutin preparation, and Comstock Foods, Alton, NY, for the apple peels. Accepted for publication 16 August 1988. ABSTRACT K61ler, W., and Parker, D. M. 1989. Purification and characterization of cutinase from Venturia inaequalis. Phytopathology 79:278-283. Venturia inaequalis was grown in a culture medium containing purified cutinase from V. inaequalisis optimal at a pH of 6 and thus different from apple cutin as the sole carbon source. After 8 wk of growth an esterase was the alkaline pH-optimum reported for other purified cutinases. The isolated from the culture fluid and purified to apparent homogeneity. The hydrolysis of the model esterp-nitrophenyl butyrate was less affected by the enzyme hydrolyzed tritiated cutin and thus was identified as cutinase. The pH. The esterase activity was strongly inhibited by diisopropyl purified cutinase is a glycoprotein with a molecular mass of 21-23 kg/ mol, fluorophosphate, and the phosphorylation of one serine was sufficient for as determined by various procedures. Remarkable structural features are a complete inhibition. Thus, cutinase from V. inaequalis belongs to the class high content of glycine, a high content of nonpolar amino acids, two of serine hydrolases, a characterisitic shared with other fungal cutinases. disulfide bridges, and a high degree of hydrophobicity. -

High Diversity and Morphological Convergence Among Melanised Fungi from Rock Formations in the Central Mountain System of Spain
Persoonia 21, 2008: 93–110 www.persoonia.org RESEARCH ARTICLE doi:10.3767/003158508X371379 High diversity and morphological convergence among melanised fungi from rock formations in the Central Mountain System of Spain C. Ruibal1, G. Platas2, G.F. Bills2 Key words Abstract Melanised fungi were isolated from rock surfaces in the Central Mountain System of Spain. Two hundred sixty six isolates were recovered from four geologically and topographically distinct sites. Microsatellite-primed biodiversity PCR techniques were used to group isolates into genotypes assumed to represent species. One hundred and sixty black fungi three genotypes were characterised from the four sites. Only five genotypes were common to two or more sites. Capnodiales Morphological and molecular data were used to characterise and identify representative strains, but morphology Chaetothyriales rarely provided a definitive identification due to the scarce differentiation of the fungal structures or the apparent Dothideomycetes novelty of the isolates. Vegetative states of fungi prevailed in culture and in many cases could not be reliably dis- extremotolerance tinguished without sequence data. Morphological characters that were widespread among the isolates included scarce micronematous conidial states, endoconidia, mycelia with dark olive-green or black hyphae, and mycelia with torulose, isodiametric or moniliform hyphae whose cells develop one or more transverse and/or oblique septa. In many of the strains, mature hyphae disarticulated, suggesting asexual reproduction by a thallic micronematous conidiogenesis or by simple fragmentation. Sequencing of the internal transcribed spacers (ITS1, ITS2) and 5.8S rDNA gene were employed to investigate the phylogenetic affinities of the isolates. According to ITS sequence alignments, the majority of the isolates could be grouped among four main orders of Pezizomycotina: Pleosporales, Dothideales, Capnodiales, and Chaetothyriales. -

Letters to Nature
letters to nature Received 7 July; accepted 21 September 1998. 26. Tronrud, D. E. Conjugate-direction minimization: an improved method for the re®nement of macromolecules. Acta Crystallogr. A 48, 912±916 (1992). 1. Dalbey, R. E., Lively, M. O., Bron, S. & van Dijl, J. M. The chemistry and enzymology of the type 1 27. Wolfe, P. B., Wickner, W. & Goodman, J. M. Sequence of the leader peptidase gene of Escherichia coli signal peptidases. Protein Sci. 6, 1129±1138 (1997). and the orientation of leader peptidase in the bacterial envelope. J. Biol. Chem. 258, 12073±12080 2. Kuo, D. W. et al. Escherichia coli leader peptidase: production of an active form lacking a requirement (1983). for detergent and development of peptide substrates. Arch. Biochem. Biophys. 303, 274±280 (1993). 28. Kraulis, P.G. Molscript: a program to produce both detailed and schematic plots of protein structures. 3. Tschantz, W. R. et al. Characterization of a soluble, catalytically active form of Escherichia coli leader J. Appl. Crystallogr. 24, 946±950 (1991). peptidase: requirement of detergent or phospholipid for optimal activity. Biochemistry 34, 3935±3941 29. Nicholls, A., Sharp, K. A. & Honig, B. Protein folding and association: insights from the interfacial and (1995). the thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281±296 (1991). 4. Allsop, A. E. et al.inAnti-Infectives, Recent Advances in Chemistry and Structure-Activity Relationships 30. Meritt, E. A. & Bacon, D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 277, 505± (eds Bently, P. H. & O'Hanlon, P. J.) 61±72 (R. Soc. Chem., Cambridge, 1997).