GREENWALD-DISSERTATION.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evaluating Feruloyl Esterase—Xylanase Synergism For
agronomy Article Evaluating Feruloyl Esterase—Xylanase Synergism for Hydroxycinnamic Acid and Xylo-Oligosaccharide Production from Untreated, Hydrothermally Pre-Treated and Dilute-Acid Pre-Treated Corn Cobs Lithalethu Mkabayi 1 , Samkelo Malgas 1 , Brendan S. Wilhelmi 2 and Brett I. Pletschke 1,* 1 Enzyme Science Programme (ESP), Department of Biochemistry and Microbiology, Rhodes University, Grahamstown, Eastern Cape 6140, South Africa; [email protected] (L.M.); [email protected] (S.M.) 2 Department of Biochemistry and Microbiology, Rhodes University, Grahamstown, Eastern Cape 6140, South Africa; [email protected] * Correspondence: [email protected]; Tel.: +27-46-6038081 Received: 4 April 2020; Accepted: 30 April 2020; Published: 13 May 2020 Abstract: Agricultural residues are considered the most promising option as a renewable feedstock for biofuel and high valued-added chemical production due to their availability and low cost. The efficient enzymatic hydrolysis of agricultural residues into value-added products such as sugars and hydroxycinnamic acids is a challenge because of the recalcitrant properties of the native biomass. Development of synergistic enzyme cocktails is required to overcome biomass residue recalcitrance, and achieve high yields of potential value-added products. In this study, the synergistic action of two termite metagenome-derived feruloyl esterases (FAE5 and FAE6), and an endo-xylanase (Xyn11) from Thermomyces lanuginosus, was optimized using 0.5% (w/v) insoluble wheat arabinoxylan (a model substrate) and then applied to 1% (w/v) corn cobs for the efficient production of xylo-oligosaccharides (XOS) and hydroxycinnamic acids. The enzyme combination of 66% Xyn11 and 33% FAE5 or FAE6 (protein loading) produced the highest amounts of XOS, ferulic acid, and p-coumaric acid from untreated, hydrothermal, and acid pre-treated corn cobs. -

Feruloyl Esterases: Biocatalysts to Overcome Biomass Recalcitrance and for the Production of Bioactive Compounds Dyoni M
Feruloyl esterases: Biocatalysts to overcome biomass recalcitrance and for the production of bioactive compounds Dyoni M. Oliveira, Thatiane R. Mota, Bianca Oliva, Fernando Segato, Rogério Marchiosi, Osvaldo Ferrarese-Filho, Craig Faulds, Wanderley D. dos Santos To cite this version: Dyoni M. Oliveira, Thatiane R. Mota, Bianca Oliva, Fernando Segato, Rogério Marchiosi, et al.. Feru- loyl esterases: Biocatalysts to overcome biomass recalcitrance and for the production of bioactive com- pounds. Bioresource Technology, Elsevier, 2019, 278, pp.408-423. 10.1016/j.biortech.2019.01.064. hal-02627378 HAL Id: hal-02627378 https://hal.inrae.fr/hal-02627378 Submitted on 26 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Accepted Manuscript Review Feruloyl esterases: Biocatalysts to overcome biomass recalcitrance and for the production of bioactive compounds Dyoni M. Oliveira, Thatiane R. Mota, Bianca Oliva, Fernando Segato, Rogério Marchiosi, Osvaldo Ferrarese-Filho, Craig -

Transcriptomic Analysis of Calonectria Pseudoreteaudii During Various Stages of Eucalyptus Infection
RESEARCH ARTICLE Transcriptomic Analysis of Calonectria pseudoreteaudii during Various Stages of Eucalyptus Infection Xiaozhen Ye1,2,3, Hongyi Liu1,3, Yajie Jin1,3, Mengmeng Guo1,3, Aizhen Huang1,3, Quanzhu Chen2,3, Wenshuo Guo1,3, Feiping Zhang1, Lizhen Feng1,3* 1 Forestry College, Fujian Agriculture and Forestry University, Fuzhou, China, 2 Jinshan College, Fujian Agriculture and Forestry University, Fuzhou, China, 3 Institute of Forestry Protection, Fujian Agriculture and Forestry University, Fuzhou, China a1111111111 a1111111111 * [email protected] a1111111111 a1111111111 a1111111111 Abstract Eucalyptus leaf blight caused by Calonectria spp. is a serious disease in Eucalyptus seed- ling and plantations. However, the molecular mechanisms of the infection process and path- OPEN ACCESS ogenesis of Calonectria to Eucalyptus is not well-studied. In this study, we analyzed the Citation: Ye X, Liu H, Jin Y, Guo M, Huang A, Chen transcriptomes of C. pseudoreteaudii at three stages of Eucalyptus leaf infection, and in Q, et al. (2017) Transcriptomic Analysis of mycelium grown in potato dextrose broth using Illumina RNA-Seq technology. We identified Calonectria pseudoreteaudii during Various Stages 161 differentially expressed genes between C. pseudoreteaudii from leaf and mycelium of Eucalyptus Infection. PLoS ONE 12(1): grown in potato dextrose broth. GO and KEGG enrichment analyses of these genes sug- e0169598. doi:10.1371/journal.pone.0169598 gested that they were mainly involved in oxidoreductase activity, hydrolase activity, and Editor: Shihui Yang, National Renewable Energy transmembrane transporter activity. Most of the differentially expressed genes at the early Laboratory, UNITED STATES infection stage were upregulated. These upregulated genes were mainly involved in cell wall Received: July 31, 2016 hydrolysis and toxin synthesis, suggesting a role for toxin and cell wall hydrolases in the Accepted: December 19, 2016 establishment of Calonectria leaf blight. -

The Structure of the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum Provides Insights Into Substrate Recognition
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Structure, Vol. 9, 1183–1190, December, 2001, 2001 Elsevier Science Ltd. All rights reserved. PII S0969-2126(01)00684-0 The Structure of the Feruloyl Esterase Module of Xylanase 10B from Clostridium thermocellum Provides Insights into Substrate Recognition Jose´ A.M. Prates,1,4 Nicolas Tarbouriech,2,4 molecular architectures that enhance their biological ef- Simon J. Charnock,2 Carlos M.G.A. Fontes,1 ficiency [1]. These proteins generally contain a mixture Luı´s M. A. Ferreira,1 and Gideon J. Davies2,3 of structurally discrete catalytic and noncatalytic mod- 1 Centro Interdisciplinar de Investigac¸ a˜ oem ules, which function cooperatively. This complexity re- Sanidade Animal flects the intricate and recalcitrant nature of their substrate, Faculdade de Medicina Veteterina´ ria the plant cell wall, a conglomerate that comprises cellu- Po´ lo Universita´ rio do Alto da Ajuda lose, hemicelluloses, and lignin, and is further fortified 1300-477 Lisboa Codex through the establishment of covalent crosslinkages. Portugal Xylan is the most abundant and important hemicellu- 2 Structural Biology Laboratory lose. It is comprised of a backbone of -1,4-linked Department of Chemistry D-xylopyranose units decorated, at O2 and/or O3, with University of York L-arabinofuranose, acetate or 4-O-methyl glucuronic acid Heslington, York YO10 5DD moieties (Figure 1). Linkage between xylan and lignin, two United Kingdom major plant cell wall components, is established by cin- namic acids, which bridge the arabinofuranosyl side chains of the former to the latter. -

Ferulic Acid Production from Banana Stem Waste: Kinetic Study
FERULIC ACID PRODUCTION FROM BANANA STEM WASTE: KINETIC STUDY JOANNE TEY CHOO HONG Thesis submitted in partial fulfilment of the requirements for the award of the degree of Bachelor of Chemical Engineering (Biotechnology) Faculty of Chemical & Natural Resources Engineering UNIVERSITI MALAYSIA PAHANG DECEMBER 2013 ©JOANNE TEY CHOO HONG (2013) III ABSTRACT For an agriculture-intensive country like Malaysia, the abundantly-available organic wastes such as banana stem provide an opportunity for the conversion into highly- sought compounds such as ferulic acid. Ferulic acid offered variety of physiological benefits used in cosmetics, food preservation and bio-production of vanillin. In this research, the soil mixed culture was mixed with banana stem wastes in an incubator shaker followed by sample collection at every 6-hours interval for the next 60 hours. Next, HPLC (high performance liquid chromatography) analysis was performed to determine the amount of ferulic acid presence. The purpose of this study was to determine the kinetic constants (Kx, Km and Vmax) from the modified biomass and Michaelis –Menten equation by using the Runge-Kutta 4th method with the aids of Microsoft Excel Solver. The kinetic constants were reported to be Kx = 0.005/ hour, Km = 0.044 mmol/ L and Vmax = 3.66 x10 -6 μmol/ min. The yield for ferulic acid production was 14.54 mg/g. Furthermore, the R2 for biomass and substrate concentration were 0.86 and 0.89 respectively. In this research, the kinetic constants and the yield for ferulic acid production were taking precedent compared to other researches due to the banana stem waste used was high in nutrient content and easy to access by soil mixed cultures. -

(12) United States Patent (10) Patent No.: US 6,365,390 B1 Blum Et Al
USOO6365390B1 (12) United States Patent (10) Patent No.: US 6,365,390 B1 Blum et al. (45) Date of Patent: Apr. 2, 2002 (54) PHENOLIC ACID ESTERASES, CODING Chen et al. (1995) “A Cyclophilin from the Polycentric SEQUENCES AND METHODS Anaerobic Rumen Fungus Orpinomyces sp. Strain PC-2 is Highly Homologous to Vertebrate Cyclophilin B” Proc. (75) Inventors: David L. Blum, San Diego, CA (US); Natl. Acad. Sci. USA 92:2587-2591.* Irina Kataeva, Athens, GA (US); Chritov and Prior (1993) “Esterases of Xylan-Degrading Xin-Liang Li, Athens, GA (US); Lars Microorganisms: Production, Properties, and Significance' G. Ljungdahl, Athens, GA (US) Enzyme Microb. Technol. 15:460-475.* Dalrymple and Swadling (1997) “Expression of a Butyriv (73) Assignee: University of Georgia Research ibrio fibrisolvens E14 Gene (cinB) Encoding an Enzyme Foundation, Inc., Athens, GA (US) with Cinnamoyl Ester Hydrolase Activity is Negatively Regulated by the Product of an Adjacent Gene (cinR)” (*) Notice: Subject to any disclaimer, the term of this Microbiology 143:1203–1210. patent is extended or adjusted under 35 Dalrymple et al. (1996) “Cloning of a Gene Encoding U.S.C. 154(b) by 0 days. Cinnamoyl Ester Hydrolase from the Ruminal Bacterium Butyrivibrio fibrisolvens E14 by a Novel Method” FEMS (21) Appl. No.: 09/390,234 Microbiology Letters 143:115-120. De Vries et al. (1997) “The faeA Genes from Aspergillus (22) Filed: Sep. 3, 1999 niger and Aspergillus tubingensis Encode Ferulic Acid Related U.S. Application Data Esterases Involved in Degradation of Complex Cell Wall (60) Provisional application No. 60/099,136, filed on Sep. 4, Polysaccharides' Applied and Environmental Microbiology 1998. -

BMC Genomics 2014, 15:535
Zuleta et al. BMC Genomics 2014, 15:535 http://www.biomedcentral.com/1471-2164/15/535 RESEARCH ARTICLE Open Access The complete genome of Burkholderia phenoliruptrix strain BR3459a, a symbiont of Mimosa flocculosa: highlighting the coexistence of symbiotic and pathogenic genes Luiz Fernando Goda Zuleta1†, Claúdio de Oliveira Cunha2,3†, Fabíola Marques de Carvalho1†, Luciane Prioli Ciapina1, Rangel Celso Souza1, Fábio Martins Mercante4, Sergio Miana de Faria5, José Ivo Baldani5, Rosangela Straliotto5, Mariangela Hungria6 and Ana Tereza Ribeiro de Vasconcelos1* Abstract Background: Burkholderia species play an important ecological role related to xenobiosis, the promotion of plant growth, the biocontrol of agricultural diseases, and symbiotic and non-symbiotic biological nitrogen fixation. Here, we highlight our study as providing the first complete genome of a symbiotic strain of B. phenoliruptrix, BR3459a (=CLA1), which was originally isolated in Brazil from nodules of Mimosa flocculosa and is effective in fixing nitrogen in association with this leguminous species. Results: Genomic comparisons with other pathogenic and non-pathogenic Burkholderia strains grouped B. phenoliruptrix BR3459a with plant-associated beneficial and environmental species, although it shares a high percentage of its gene repertoire with species of the B. cepacia complex (Bcc) and "pseudomallei" group. The genomic analyses showed that the bce genes involved in exopolysaccharide production are clustered together in the same genomic region, constituting part of the Group III cluster of non-pathogenic bacteria. Regarding environmental stresses, we highlight genes that might be relevant in responses to osmotic, heat, cold and general stresses. Furthermore, a number of particularly interesting genes involved in the machinery of the T1SS, T2SS, T3SS, T4ASS and T6SS secretion systems were identified. -
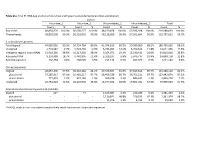
Table S1a. Trial #1 RNA-Seq Analysis of Virus-Free and Hypovirus
Table S1a. Trial #1 RNA-Seq analysis of virus-free and hypovirus-transfected Sclerotinia sclerotiorum Library Virus-free_1 Virus-free_2 Virus-infected_1 Virus-infected_2 Total Count % Count % Count % Count % Count % Raw reads 48,854,170 100.0% 56,568,277 100.0% 48,169,458 100.0% 57,396,928 100.0% 210,988,833 100.0% Filtered reads 48,810,008 99.9% 56,518,010 99.9% 48,118,009 99.9% 57,341,604 99.9% 210,787,631 99.9% S. sclerotiorum genome Total aligned 44,085,026 90.3% 50,724,764 89.8% 41,374,916 86.0% 50,580,960 88.2% 186,765,666 88.6% Unaligned 4,724,982 9.7% 5,793,246 10.3% 6,743,093 14.0% 6,760,644 11.8% 24,021,965 11.5% Intergenic regions (non-rRNA) 13,918,285 28.5% 16,317,039 28.9% 9,354,071 19.4% 15,230,421 26.6% 54,819,816 25.8% Ribosomal RNA 9,150,409 18.7% 6,442,905 11.4% 3,329,107 6.8% 5,976,737 10.4% 24,899,158 11.8% Retrotransposons 202,264 0.4% 268,505 0.5% 251,134 0.5% 499,377 0.9% 1,221,280 0.6% Coding sequences Aligned 28,057,390 57.5% 36,310,362 64.2% 29,370,507 61.0% 37,649,843 65.7% 131,388,102 62.1% plus strand 27,285,917 97.3% 35,481,217 97.7% 28,409,529 96.7% 36,766,211 97.7% 127,942,874 97.3% minus strand 771,473 2.7% 829,145 2.3% 960,978 3.3% 883,632 2.3% 3,445,228 2.7% Unaligned 20,752,618 42.5% 20,207,648 35.8% 18,747,502 39.0% 19,691,761 34.3% 79,399,529 37.9% Sclerotinia sclerotiorum hypovirus 2L (SsHV2L) Aligned 62* - 77 - 1,165,087 2.4% 216,286 0.4% 1,381,450 1.4% plus strand - - - - 1,150,841 98.8% 210,533 97.3% 1,361,374 98.1% minus strand - - - - 14,246 1.2% 5,753 2.7% 19,999 1.9% *SsHV2L reads in non-inoculated samples likely result from errors in barcode sequences. -

(12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel Et Al
USOO8561811 B2 (12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel et al. (45) Date of Patent: Oct. 22, 2013 (54) SUBSTRATE FOR IMMOBILIZING (56) References Cited FUNCTIONAL SUBSTANCES AND METHOD FOR PREPARING THE SAME U.S. PATENT DOCUMENTS 3,952,053 A 4, 1976 Brown, Jr. et al. (71) Applicants: Christian Gert Bluchel, Singapore 4.415,663 A 1 1/1983 Symon et al. (SG); Yanmei Wang, Singapore (SG) 4,576,928 A 3, 1986 Tani et al. 4.915,839 A 4, 1990 Marinaccio et al. (72) Inventors: Christian Gert Bluchel, Singapore 6,946,527 B2 9, 2005 Lemke et al. (SG); Yanmei Wang, Singapore (SG) FOREIGN PATENT DOCUMENTS (73) Assignee: Temasek Polytechnic, Singapore (SG) CN 101596422 A 12/2009 JP 2253813 A 10, 1990 (*) Notice: Subject to any disclaimer, the term of this JP 2258006 A 10, 1990 patent is extended or adjusted under 35 WO O2O2585 A2 1, 2002 U.S.C. 154(b) by 0 days. OTHER PUBLICATIONS (21) Appl. No.: 13/837,254 Inaternational Search Report for PCT/SG2011/000069 mailing date (22) Filed: Mar 15, 2013 of Apr. 12, 2011. Suen, Shing-Yi, et al. “Comparison of Ligand Density and Protein (65) Prior Publication Data Adsorption on Dye Affinity Membranes Using Difference Spacer Arms'. Separation Science and Technology, 35:1 (2000), pp. 69-87. US 2013/0210111A1 Aug. 15, 2013 Related U.S. Application Data Primary Examiner — Chester Barry (62) Division of application No. 13/580,055, filed as (74) Attorney, Agent, or Firm — Cantor Colburn LLP application No. -

Bioprospecting for Microorganisms and Enzymes with Biorefining Potential
Bioprospecting for Microorganisms and Enzymes with Biorefining Potential by Laura Frances Lyons Supervisors Dr Kerrie Farrar and Dr Justin Pachebat A thesis submitted at the Institute of Biological, Environmental and Rural Sciences (Aberystwyth University), for the degree of Doctor of Philosophy. 2015 1 2 Declaration This work has not previously been accepted in substance for any degree and is not being concurrently submitted in candidature for any degree. Signed ...................................................................... (candidate) Date ........................................................................ STATEMENT 1 This thesis is the result of my own investigations, except where otherwise stated. Where *correction services have been used, the extent and nature of the correction is clearly marked in a footnote(s). Other sources are acknowledged by footnotes giving explicit references. A bibliography is appended. Signed ..................................................................... (candidate) Date ........................................................................ [*this refers to the extent to which the text has been corrected by others] STATEMENT 2 I hereby give consent for my thesis, if accepted, to be available for photocopying and for inter-library loan, and for the title and summary to be made available to outside organisations. Signed ..................................................................... (candidate) Date ........................................................................ Word count -

ABSTRACT SHU, XIAOMEI. Pathogenesis and Host Response
ABSTRACT SHU, XIAOMEI. Pathogenesis and Host Response During Infection of Maize Kernels by Aspergillus flavus and Fusarium verticillioides. (Under the direction of Dr. Gary A. Payne.) Developing maize kernels are vulnerable to colonization by microbes. When colonization allows proliferation of the microbe at the expense of the host, disease occurs. The ascomycete fungal pathogens Aspergillus flavus and Fusarium verticillioides are capable of colonizing maize kernels, causing ear rots and contamination of the kernel with mycotoxins. These diseases lead to significant losses of crop yield and quality, and constitute a threat to food safety and human health. Thus, the significance of these diseases has prompted extensive research efforts to understand these plant-parasite interactions. However, pathogenesis and resistance mechanisms remain poorly characterized, hampering the development of effective control strategies. No commercial maize lines are completely resistant to these fungi. We applied an integrated approach consisting of histology, in situ gene expression and transcriptional profiling to better understand the nature of the interactions that occur between maize kernels and these fungi. Maize inbred line B73 was hand pollinated and inoculated with either A. flavus or F. verticillioides by wounding the kernel with a needle bearing conidia. Histological staining of the kernel sections revealed fungal mycelium in kernels adjacent to the inoculation site by 48 hours post inoculation (hpi), and in all tissues at 96 hpi. Compared with F. verticillioides, A. flavus more aggressively colonized kernel tissue and formed a unique biofilm-like structure around the scutellum. Transcriptome profiling using RNA-sequencing (RNA-seq) coupled with pathway analysis showed that these fungi were recognized by the kernel tissues prior to visible colonization. -

Characterization and Engineering of a Two-Enzyme System for Plastics Depolymerization
Characterization and engineering of a two-enzyme system for plastics depolymerization Brandon C. Knotta,1, Erika Ericksona,1, Mark D. Allenb,1, Japheth E. Gadoa,c,1, Rosie Grahamb, Fiona L. Kearnsd, Isabel Pardoa, Ece Topuzlua,e, Jared J. Andersona, Harry P. Austinb, Graham Dominicka, Christopher W. Johnsona, Nicholas A. Rorrera, Caralyn J. Szostkiewicza, Valérie Copiée, Christina M. Paynec, H. Lee Woodcockd, Bryon S. Donohoef, Gregg T. Beckhama,2, and John E. McGeehanb,2 aRenewable Resources and Enabling Sciences Center, National Renewable Energy Laboratory, Golden, CO 80401; bCentre for Enzyme Innovation, School of Biological Sciences, Institute of Biological and Biomedical Sciences, University of Portsmouth, Portsmouth PO1 2DY, United Kingdom; cDepartment of Chemical and Materials Engineering, University of Kentucky, Lexington, KY 40506; dDepartment of Chemistry, University of South Florida, Tampa, FL 33620; eDepartment of Chemistry and Biochemistry, Montana State University, Bozeman, MT 59717; and fBiosciences Center, National Renewable Energy Laboratory, Golden, CO 80401 Edited by Alexis T. Bell, University of California, Berkeley, CA, and approved August 26, 2020 (received for review April 11, 2020) Plastics pollution represents a global environmental crisis. In re- glycol (EG). Given the prevalence of esterase enzymes in nature, sponse, microbes are evolving the capacity to utilize synthetic PET biodegradation has been studied for nearly two decades, with polymers as carbon and energy sources. Recently, Ideonella sakaien- multiple cutinase enzymes reported to perform depolymerization sis wasreportedtosecreteatwo-enzyme system to deconstruct (17–26). In 2016, Yoshida et al. (10) reported the discovery and polyethylene terephthalate (PET) to its constituent monomers. Spe- characterization of the soil bacterium, Ideonella sakaiensis 201-F6, cifically, the I.