Variability Analysis of Some Genotypes in Nigeria Tea (Camellia Sinensis) Germplasm
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Preliminary Catalogue of Isaac Roberts's Collection of Photographs of Celestial Objects
ASTRONOMISCHE NACHRICHTEN. Nr. 4154. Band 174. 2. Preliminary catalogue of Isaac Roberts’s collection of photographs of celestial objects. By Dorotha Isaac-Roberts. The Isaac Roberts collection of celestial photographs The number of the copies of the forthcoming paper comprises 2485 original negatives of stars, star-clusters, being very limited, the observatories and astronomers, official nebulae and other celestial objects together with many posi. or amateur, who are specially interested in photographic tives on glass and on paper, taken by the late Dr. Isaac astronomy will please send in their names to the address Roberts or, under his incessant supervision, by his assistant given below, early in 1907, in order that the various parts W. S. Franks, F. R. A. S., at Dr. Roberts’s Private Obser- of the complete catalogue may be sent to them in course vatory, Kennessee, Maghull, near Liverpool (1885-1 890) and, of time. from 1890 to 1904, at Starfield, Crowborough, Sussex. Positives- on-glass reproduced from the Isaac Roberts Of the above mentioned negatives, negatives will be lent for the purpose of micrometric mea- I) 1412 were taken with the zo-in Reflector (focal surements, if, application be made and provided that the length = 98.0 ins) and with an exposure of the plate of documents be returned after completion of the measurements. ninety minutes in general. In the preliminary catalogue given below, the numbers 2) 648 with the 5-in Cooke lens (f. 1. = 19.22 ins) of Dr. Dreyer’s New General Catalogue of Nebulae and which was mounted on the tube of the zo-in Reflector; or, Clusters of Stars and of Dr. -

Cetus - the Whale
May 18 2021 Cetus - The Whale Observed: No Object Her Type Mag Alias/Notes IC 5384 Non-Existent NGC 7813 MCG -2-1-16 MK 936 IRAS 15-1215 PGC 287 IC 1528 Non-Existent NGC 7826 H29-8 Non-Existent Asterism IC 1533 Non-Existent NGC 34 Non-Existent NGC 17 NGC 58 Non-Existent NGC 47 PGC 967 MCG -1-1-55 IRAS 119-726 NGC 54 Glxy SB(r)a? 14.6 MCG -1-1-60 PGC 1011 NGC 59 Glxy SA(rs)0-: 13.1 ESO 539-4 MCG -4-1-26 PGC 1034 NGC 62 Glxy (R)SB(r)a: 12.3 MCG -2-1-43 IRAS 145-1345 PGC 1125 NGC 64 Glxy SB(s)bc 14 MCG -1-1-68 IRAS 149-706 PGC 1149 IC 5 Glxy E 14.8 MCG -2-1-47 IRAS 148-951 PGC 1145 NGC 73 Glxy SAB(rs)bc: 13.5 MCG -3-1-26 PGC 1211 NGC 65 Glxy SAB(rs)0-: 14.4 ESO 473-10A MCG -4-2-1 PGC 1229 NGC 66 Glxy SB(r)b pec 14.2 ESO 473-10 MCG -4-2-2 IRAS 165-2312 PGC 1236 IC 9 Glxy Sb(r) 16.1 MCG -2-2-1 IRAS 171-1423 PGC 1271 NGC 77 Glxy SA0-: 15.7 ESO 473-15 PGC 1290 NGC 102 Glxy S0/a 14.4 MCG -2-2-11 PGC 1542 NGC 107 Glxy Sbc 14.6 MCG -2-2-14 PGC 1606 NGC 111 Non-Existent NGC 113 Glxy SA0-: 13.5 MCG -1-2-16 PGC 1656 NGC 114 Glxy SB(rs)0: 14.7 UGC 259 MCG 0-2-27 MK 946 CGCG 383-14 KUG 24-20A PGC 1660 NGC 116 Non-Existent MCG -1-2-17 PGC 1671 NGC 117 Glxy S0+: sp 15.3 MCG 0-2-29 CGCG 383-15 PGC 1674 NGC 118 Glxy I0? 14.8 UGC 264 MCG 0-2-32 MK 947 CGCG 383-16 UM244 3ZW9 IRAS 247-203 PGC 1678 NGC 120 Glxy SB0^: 14.4 UGC 267 MCG 0-2-33 CGCG 383-17 PGC 1693 NGC 122 Non-Existent NGC 123 Non-Existent NGC 124 Glxy SA(s)c 13.7 UGC 271 MCG 0-2-38 CGCG 383-18 IRAS 253-205 PGC 1715 IC 15 Non-Existent IC 16 Glxy E? 14.7 MCG -2-2-17 IRAS 255-1322 PGC 1730 IC 17 -

Alma Observations of Warm Molecular Gas and Cold Dust in Ngc 34∗
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Caltech Authors The Astrophysical Journal, 787:48 (10pp), 2014 May 20 doi:10.1088/0004-637X/787/1/48 C 2014. The American Astronomical Society. All rights reserved. Printed in the U.S.A. ALMA OBSERVATIONS OF WARM MOLECULAR GAS AND COLD DUST IN NGC 34∗ C. K. Xu1,C.Cao1,2,3,N.Lu1,Y.Gao4, P. van der Werf5,A.S.Evans6,7, J. M. Mazzarella1, J. Chu8,S.Haan9, T. Diaz-Santos1, R. Meijerink5, Y.-H. Zhao1,4, P. Appleton1, L. Armus1, V. Charmandaris10,11,12,S.Lord1, E. J. Murphy1, D. B. Sanders8, B. Schulz1, and S. Stierwalt6,13 1 Infrared Processing and Analysis Center, MS 100-22, California Institute of Technology, Pasadena, CA 91125, USA 2 School of Space Science and Physics, Shandong University at Weihai, Weihai, Shandong 264209, China; [email protected] 3 Shandong Provincial Key Laboratory of Optical Astronomy and Solar-Terrestrial Environment, Weihai, Shandong 264209, China 4 Purple Mountain Observatory, Chinese Academy of Sciences, 2 West Beijing Road, Nanjing 210008, China 5 Leiden Observatory, Leiden University, P.O. Box 9513, NL-2300 RA Leiden, Netherlands 6 Department of Astronomy, University of Virginia, P.O. Box 400325, Charlottesville, VA 22904, USA 7 National Radio Astronomy Observatory, Charlottesville, VA 22904, USA 8 Institute for Astronomy, University of Hawaii, 2680 Woodlawn Drive, Honolulu, HI 96816, USA 9 CSIRO Astronomy and Space Science, ATNF, P.O. Box 76, Epping 1710, Australia 10 Department of Physics, University of Crete, GR-71003, Heraklion, Greece 11 Institute for Astronomy, Astrophysics, Space Applications & Remote Sensing, National Observatory of Athens, GR-15236, Penteli, Greece 12 Chercheur Associe,´ Observatoire de Paris, F-75014, Paris, France 13 Spitzer Science Center, California Institute of Technology, 1200 E. -

Ngc Catalogue Ngc Catalogue
NGC CATALOGUE NGC CATALOGUE 1 NGC CATALOGUE Object # Common Name Type Constellation Magnitude RA Dec NGC 1 - Galaxy Pegasus 12.9 00:07:16 27:42:32 NGC 2 - Galaxy Pegasus 14.2 00:07:17 27:40:43 NGC 3 - Galaxy Pisces 13.3 00:07:17 08:18:05 NGC 4 - Galaxy Pisces 15.8 00:07:24 08:22:26 NGC 5 - Galaxy Andromeda 13.3 00:07:49 35:21:46 NGC 6 NGC 20 Galaxy Andromeda 13.1 00:09:33 33:18:32 NGC 7 - Galaxy Sculptor 13.9 00:08:21 -29:54:59 NGC 8 - Double Star Pegasus - 00:08:45 23:50:19 NGC 9 - Galaxy Pegasus 13.5 00:08:54 23:49:04 NGC 10 - Galaxy Sculptor 12.5 00:08:34 -33:51:28 NGC 11 - Galaxy Andromeda 13.7 00:08:42 37:26:53 NGC 12 - Galaxy Pisces 13.1 00:08:45 04:36:44 NGC 13 - Galaxy Andromeda 13.2 00:08:48 33:25:59 NGC 14 - Galaxy Pegasus 12.1 00:08:46 15:48:57 NGC 15 - Galaxy Pegasus 13.8 00:09:02 21:37:30 NGC 16 - Galaxy Pegasus 12.0 00:09:04 27:43:48 NGC 17 NGC 34 Galaxy Cetus 14.4 00:11:07 -12:06:28 NGC 18 - Double Star Pegasus - 00:09:23 27:43:56 NGC 19 - Galaxy Andromeda 13.3 00:10:41 32:58:58 NGC 20 See NGC 6 Galaxy Andromeda 13.1 00:09:33 33:18:32 NGC 21 NGC 29 Galaxy Andromeda 12.7 00:10:47 33:21:07 NGC 22 - Galaxy Pegasus 13.6 00:09:48 27:49:58 NGC 23 - Galaxy Pegasus 12.0 00:09:53 25:55:26 NGC 24 - Galaxy Sculptor 11.6 00:09:56 -24:57:52 NGC 25 - Galaxy Phoenix 13.0 00:09:59 -57:01:13 NGC 26 - Galaxy Pegasus 12.9 00:10:26 25:49:56 NGC 27 - Galaxy Andromeda 13.5 00:10:33 28:59:49 NGC 28 - Galaxy Phoenix 13.8 00:10:25 -56:59:20 NGC 29 See NGC 21 Galaxy Andromeda 12.7 00:10:47 33:21:07 NGC 30 - Double Star Pegasus - 00:10:51 21:58:39 -

Evaluation of the National Guideline Clearinghouse (NGC)
Final Report Evaluation of AHRQ’s National Guideline Clearinghouse™ (NGC) Agency for Healthcare Research and Quality August 31, 2011 AFYA, Inc. 8101 Sandy Spring Road, Third Floor Laurel, MD 20707 Final Contract Report: NGC Evaluation Final Submitted: 9/29/2011 Final Contract Report Evaluation of AHRQ’s National Guideline Clearinghouse™ (NGC) Prepared for: Agency for Healthcare Research and Quality U.S. Department of Health and Human Services 540 Gaither Road Rockville, MD 20850 www.ahrq.gov Prepared by: AFYA, Inc. Analytical and Technical Support Division 8101 Sandy Spring Road, Third Floor Laurel, MD 20707 Authors: Michelle Bieber-Tregear AFYA, Inc. Jenice James AFYA, Inc. Debra Dekker AFYA, Inc. Craig Dearfield AFYA, Inc. Jaclyn Marshall Lewin Group Jacob Epstein Lewin Group Carol Simon Lewin Group Financial Declarations None of the authors has any affiliations or financial involvement that conflict with the material presented in this report. Policy Statement This report was prepared by AFYA, Inc. and The Lewin Group under Contract No. 4203; Order No. 3, entitled “Evaluation of the National Guideline Clearinghouse™ (NGC),” with the Department of Health and Human Services’ Agency for Healthcare Research and Quality. Table of Contents List of Figures............................................................................................................................ 5 List of Tables ............................................................................................................................. 6 AHRQ Contract No. 4203; -

Selecting an Award to Write Page 1 of 7 Leadership Awards 1, 2, and 3 Are Judged by an Anonymous Committee Appointed by the FFGC President
How to Select a FFGC Award to write from your Garden Club’s Programs or Events Valerie Seinfeld Civic Awards ? Horticulture ? Community Projects/ Historical ? Flower Show Achievement Awards ? Conservation ? Leadership ? District Awards ? Tree Planting ? Publications & Social Media ? Junior Gardening ? Landscaping ? Miscellaneous Awards ? Special Achievement ? How to Select which FFGC Award to write from your Garden Club’s Programs or Events www.ffgc.org Members Tab - click to drop down and Awards is first. Check back often as new awards may be added each year. Begin by looking at the Category the award falls under. Read it in detail. Does your program or event meet the criteria? This handy document will give you an idea before you go to the computer if there is an award title that matches your program or event. Once you decide on an award to write, download the application form and fill out the top portion. Save it for later when you are ready to start. But Not ALL: Remember most awards, but not all, are due at the same time. Most are sent to the same place, but not all. Most are attention to the Awards Chair, but not all. Make note of exceptions. Reread the award description in detail. Give the Chair of the Program or Event a list of items you need to complete the Award. Ex. Pictures before and after, Budget and where the money came from. How many were involved in this award, all members, percentage of members, community members? Valerie Seinfeld Selecting an Award to Write page 1 of 7 Leadership Awards 1, 2, and 3 are judged by An Anonymous Committee appointed by the FFGC President. -

Appendix a Planning an Astrophotography Imaging Session
Appendix A Planning an Astrophotography Imaging Session One way to obtain a high percentage of successful photographs is to look at imaging a deep space object as a project. The first step of any project is to define the project’s goals and objectives followed by detailed planning and then executing the plan. Perhaps the best way to explain an astrophotography project is by an example. For our example, let’s assume a location at 38° north latitude in a suburban area with moderate levels of light pollution with a skyglow of magnitude 19 per arcsec- ond squared. The imaging kit is an Orion ST-80A 80 mm f/5 refractor, a Canon EOS T3/1100D camera with an interval timer and a Celestron 4 SE mount on a wedge. The current time of the year is early summer and we see an image of the galaxy NGC 2403 posted by someone on an internet forum, in an astronomy maga- zine, etc. The galaxy looks interesting and we decide that we would like to make our own image of it. Before getting heavily involved in planning we need answers to the following questions: • Is the galaxy visible at our viewing location? • When is the optimum time to photograph the galaxy? • Is our equipment capable of capturing a decent image of NGC 2403? The literature tells us that NGC 2403 has an apparent magnitude 8.4, an apparent size of 21.4 × 10.7 arcminutes, and is located in the Constellation Camelopardalis. It is object number 7 in the Caldwell list for small telescopes and is often observed with a pair of binoculars. -

Gear-Flex Coupling
RATHI TRANSPOWER PVT. LTD. PUNE - INDIA PRODUCT MANUAL GEAR-FLEX COUPLING TYPE – LFG/LHG R-PM-G-01/02-01/14 Page 1 INDEX CONTENTS PAGE • Standard Features 3 • At a Glance 3 • Gear-flex Family 4 • Std. Material of Construction 4 • Ratings for Standard couplings 5 • Weight & M.I. for standard couplings 6 • Customized Gear-flex Specials 7 • Half Flex Couplings with Floating Shafts 7 • Spacer Couplings 8 • Reversed Hub Combinations 8 • Lubricants 9 • Requirements of API-671 9 • Selection of Size of Coupling 10 • Typical Service Factors 11-13 • Std. Tolerances for Finish Bore & Keyway 14 • Applications 14 • Gear-flex couplings equivalent to Competitor’s couplings 15-18 R-PM-G-01/02-01/14 Page 2 GEAR-FLEX COUPLING STANDARD FEATURES OF GEAR-FLEX COUPLING ♦ Less backlash. ♦ High power to weight ratio. ♦ Compact assembly. ♦ Accommodates angular, parallel & axial misalignments. ♦ Generally used up to 120°C. Can be used for higher temperatures by using proper grade of oil or grease. ♦ Can be dynamically balanced to the required grade as per ISO-1940. AT A GLANCE SIZES Full Flexible Type LFG - 100 to 119 (20 sizes) Half Flexible Type LHG - 100 to 110 (11 sizes) RATING RANGE TYPE LFG - 5.9 kW to 17454 kW @ 100 rpm TYPE LHG- 5.9 kW to 963 kW @ 100 rpm TORQUE RANGE TYPE LFG - 559 Nm to 1666744 Nm TYPE LHG- 559 Nm to 91922 Nm BORE RANGE TYPE LFG - 13 mm. to 710 mm. TYPE LHG- 13 mm. to 260 mm. ANGULAR MISALIGNMENT - 1.5° per gear mesh Maximum COMPLIANCE WITH API-671 SPECIFICATIONS, IF REQD. -
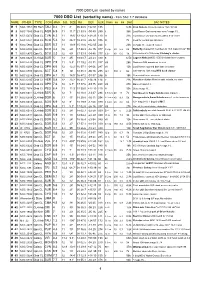
DSO List V2 Current
7000 DSO List (sorted by name) 7000 DSO List (sorted by name) - from SAC 7.7 database NAME OTHER TYPE CON MAG S.B. SIZE RA DEC U2K Class ns bs Dist SAC NOTES M 1 NGC 1952 SN Rem TAU 8.4 11 8' 05 34.5 +22 01 135 6.3k Crab Nebula; filaments;pulsar 16m;3C144 M 2 NGC 7089 Glob CL AQR 6.5 11 11.7' 21 33.5 -00 49 255 II 36k Lord Rosse-Dark area near core;* mags 13... M 3 NGC 5272 Glob CL CVN 6.3 11 18.6' 13 42.2 +28 23 110 VI 31k Lord Rosse-sev dark marks within 5' of center M 4 NGC 6121 Glob CL SCO 5.4 12 26.3' 16 23.6 -26 32 336 IX 7k Look for central bar structure M 5 NGC 5904 Glob CL SER 5.7 11 19.9' 15 18.6 +02 05 244 V 23k st mags 11...;superb cluster M 6 NGC 6405 Opn CL SCO 4.2 10 20' 17 40.3 -32 15 377 III 2 p 80 6.2 2k Butterfly cluster;51 members to 10.5 mag incl var* BM Sco M 7 NGC 6475 Opn CL SCO 3.3 12 80' 17 53.9 -34 48 377 II 2 r 80 5.6 1k 80 members to 10th mag; Ptolemy's cluster M 8 NGC 6523 CL+Neb SGR 5 13 45' 18 03.7 -24 23 339 E 6.5k Lagoon Nebula;NGC 6530 invl;dark lane crosses M 9 NGC 6333 Glob CL OPH 7.9 11 5.5' 17 19.2 -18 31 337 VIII 26k Dark neb B64 prominent to west M 10 NGC 6254 Glob CL OPH 6.6 12 12.2' 16 57.1 -04 06 247 VII 13k Lord Rosse reported dark lane in cluster M 11 NGC 6705 Opn CL SCT 5.8 9 14' 18 51.1 -06 16 295 I 2 r 500 8 6k 500 stars to 14th mag;Wild duck cluster M 12 NGC 6218 Glob CL OPH 6.1 12 14.5' 16 47.2 -01 57 246 IX 18k Somewhat loose structure M 13 NGC 6205 Glob CL HER 5.8 12 23.2' 16 41.7 +36 28 114 V 22k Hercules cluster;Messier said nebula, no stars M 14 NGC 6402 Glob CL OPH 7.6 12 6.7' 17 37.6 -03 15 248 VIII 27k Many vF stars 14.. -

My Finest NGC Album
My Finest NGC Album A detailed record of my journey through The Royal Astronomical Society of Canada’s Finest NGC list Name: ______________________________ Centre or Home Location: ______________________________ The New General Catalogue or NGC contains 7,840 entries and forms the core of most people's "life list" of observing targets. The NGC was originally published in 1888 by J.L.E. Dreyer and therefore predated photographic astronomy. The Finest NGC list, compiled by Alan Dyer complements the Messier List, as there is no overlap. The list features many fine deep-sky treasures as well as a few somewhat more challenging objects. Once you have observed all of the objects on this list, application forms can be found on the RASC website at www.rasc.ca. The FNGC certificate has been awarded since 1995. Here is an overview of the Finest NGC Observing List Finest NGC Objects Number Notes Open Clusters 12 Including the famous Double Cluster in Perseus, NGC 7789 in Cassiopeia, and NGC 6633 in Ophiuchus. Globular Clusters 2 NGC 5466 in Bootes and NGC 6712 in Scutum. Diffuse Nebulae 14 Includes the great Veil Nebula as well as the North America and Rosette nebulae. Planetary Nebulae 24 Includes many fine PN's like the Ghost of Jupiter, the Cat's Eye, the Blinking Planetary, the Helix, the Blue Snowball, and the Clown Face nebulae. Galaxies 58 Includes the amazing NGC 4565 in Coma Berenices, NGC 253 in Sculptor, and NGC 5907 in Draco. Total 110 The Finest NGC list can be started during any season. Why Record Your Observations? Recording observations is important for two reasons. -

Comparative Plant Resistance in Fifty-Five Tea Accessions, Camellia Sinensis L., in Ibadan, South West, Nigeria
Plant resistance of Camellia sinensis L ( Coleoptera: JBiopest 10(2):146-153 (2017) ComparativeChrysomelidae: plant Bruchinae) resistance in fifty-five tea accessions, Camellia sinensis L., in Ibadan, South West, Nigeria O. M.Azeez ABSTRACT Fifty five accessions comprising both local and exotic clones were assessed for their resistance to field pest at Abarakata farm, CRIN. The accessions derived from Mambilla plateau, CRIN out-station, Taraba State were screened for resistance to infestation by leaf defoliators. Results revealed that there were significant differences among the accessions in terms of the number of holes on the leaf and plant damage both in the dry and raining season. On NGC15, NGC17, NGC19, C235, C357 tea clones recorded less number of holes and total damage of leaves and thus considered highly resistant to field insect pests. Based on the rating, clones NGC 8, NGC12, NGC14, NGC24, NGC27, NGC40, NGC41, NGC42,NGC49, NGC50,NGC51, C68, C270,C318, C359, C377 were significantly resistant to the insect infestations, while NGC18, NGC22, NGC23,NGC25, NGC26, NGC29, NGC35,NGC47, NGC48, NGC53, NGC54, NGC55, C61, C136, C143, C228, C236, C327, C353, C354, C369, C370 were moderately resistant. NGC13, NGC32, NGC37, NGC38, NGC45, NGC46, C56, C74, C108, C363, C367 and C368 were the most susceptible with the highest damage indices values (P<0.05). The range of each of the resistance indices measured in the susceptible clones in both dry and wet season were number of leaf damage (43-51)(37-46), number of leaf holes (18-19)(16-20). Keywords: Clones; tea; resistance; damage; susceptible MS History: 16.10.2017 (Received)-27.11.2017 (Revised)-3.12.2017 (Accepted). -

Minterrornews.Com Special Edition 2006 the Best of Mint Error News
TM minterrornews.com Special Edition 2006 The Best of Mint Error News A Mike Byers Publication Byers Numismatic Corp mikebyers.com TM The Largest Dealer of the World’s Rarest Mint Errors U.S. & World Major Mint Errors • Die Trials • Numismatic Rarities Unique SPECIMEN Silver 1920 SL 25¢ 1838 $5 Die Trial Splasher Certificate Set of 16 Struck on Peru 20C Planchet J-A1838-6 PCGS Certified NGC MS 60 FH Unique PCGS MS 65 UNIQUE Unique 1866 $2½ Struck on a 3 Cent Pair of Indian Head 1¢ Die Caps Barber Half Nickel Planchet Obverse & Reverse Full Obverse Brockage NGC MS 66 PCGS MS 64 PCGS AU 58 UNIQUE Unique Set of Four 1921-S Morgan Dollar 1895-O Barber Dime Paraguay Gold Overstrikes Struck 45% Off-Center Obverse Die Cap NGC Certified NGC MS 63 PCGS MS 64 1846 J-110A $5 Obv Die Trial 1924 SL 25¢ 1862 Indian Head 1¢ Struck on $2½ Trial Double Struck Deep Obverse Die Cap NGC MS 65 BN ANACS AU 55 PCGS MS 62 Unique Set of Three 1887 $3 Indian Gold Proof 1942 Walking Liberty 50¢ Paraguay Gold Overstrikes Triple Struck Struck on Silver 25¢ Planchet NGC Certified PCGS PR 63 PCGS MS 65 Unique Jefferson Nickel 1802/1 $5 Draped Bust Gold 1865 2¢ Die Trial Triple Struck Obverse Deep Obverse Die Cap PCGS Certified ANACS EF 45 & Brockage 1804 $2½ Capped Bust To Right 1898 Barber 25¢ 1945-S WL 50¢ Double Struck Obverse Die Cap & Brockage Struck on El Salvador 25¢ Planchet NGC Fine 15 PCGS MS 62 NGC MS 63 UNQUE 1806 $5 Capped Bust Triple Struck 1865 $1 Indian Gold Proof 1920 Buffalo Nickel Rotated 90° Triple Struck Reverse Struck on Copper Planchet PCGS AU 50 PCGS PR 64 Cameo NGC AU 55 UNIQUE 1874 $1 U.S.