Understanding the Diversity and Community Structure of Bdelloid Rotifer Soil Communities
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(Rotifera: Monogononta) from Andhra Pradesh, India, Including Six New Distribution
Journal of Threatened Taxa | www.threatenedtaxa.org | July 2013 | 5(11): 4556–4561 A report on Lecanidae (Rotifera: Monogononta) from Andhra Pradesh, India, including six new distribution records with notes on their contemporary taxonomic ISSN Short Communication Short Online 0974-7907 nomenclature Print 0974-7893 S.Z. Siddiqi 1 & M. Karuthapandi 2 oPEN ACCESS 1,2 Zoological Survey of India, Freshwater Biological Regional Centre, Hyderabad, Andhra Pradesh 500048, India 1 [email protected] (corresponding author), 2 [email protected] Abstract: The Lecane-species complex taxonomy the world over, families. In fact the taxonomy of ubiquitous, bio- witnessed a state of flux, causing confusion and controversies, among geographically significant Lecane species-complex was world’s taxonomists over the treatment of various subgenera, taxa and sub and or infraspecific categories of the species rich genus Lecane in a state of flux for decades, and indeed a major irritant Nitzsch 1827, on the basis of structure/shape of key, morphological to taxonomists world wide, following differences, until features like foot/toes, lorica, etc. The taxonomic scenario in India, relying heavily on the classical, divergent taxonomic approaches the mid nineties; Segers (1995) finally setting to rest all presented a picture of more chaos/confusion, following poor prevailing confusions/ controversies, assigning all species accessibility to contemporary revisionary studies until the recent under a single genus Lecane Nitzsch, 1827. The Indian past. Despite revisionary -

Continued Exploration of Tanzanian Rainforests Reveals a New Echiniscid Species (Heterotardigrada)
Zoological Studies 59:18 (2020) doi:10.6620/ZS.2020.59-18 Open Access Continued Exploration of Tanzanian Rainforests Reveals a New Echiniscid Species (Heterotardigrada) Marcin Bochnak1, Katarzyna Vončina1, Reinhardt M. Kristensen2,§, and Piotr Gąsiorek1,§* 1Institute of Zoology and Biomedical Research, Jagiellonian University, Gronostajowa 9, 30-387 Kraków, Poland. *Correspondence: E-mail: [email protected] (Gąsiorek) E-mail: [email protected] (Bochnak); [email protected] (Vončina) 2Section for Biosystematics, Natural History Museum of Denmark, University of Copenhagen, Universitetsparken 15, Copenhagen Ø DK-2100, Denmark. E-mail: [email protected] (Kristensen) §RMK and PG share joint senior authorship. Received 13 January 2020 / Accepted 28 April 2020 / Published 15 June 2020 Communicated by Benny K.K. Chan The Afrotropical tardigrade fauna is insufficiently studied, and consequently its diversity in this region is severely underestimated. Ongoing sampling in the Udzungwa Mountains, Morogoro Region of Tanzania has revealed a new representative of the genus Echiniscus C.A.S. Schultze, 1840 (Echiniscidae). Echiniscus tantulus sp. nov. belongs to the spinulosus group, but it stands out from other members of this speciose Echiniscus clade by having a heteromorphic sculpture of the dorsal plates and an uncommonly stable body appendage configuration A-C-Cd-Dd-E. The new species is characteristic by being equipped with long dorsal spines and very short lateral spicules, which so far have been found only in one other species of the group, Echiniscus spinulosus (Doyère, 1840). An updated checklist of Tanzanian Echiniscidae is provided, incorporating recent advances in their classification. Key words: Biodiversity, Chaetotaxy, Cuticular sculpturing, The spinulosus group, Udzungwa Mountains. -

An Introduction to Phylum Tardigrada - Review
Volume V, Issue V, May 2016 IJLTEMAS ISSN 2278 – 2540 An Introduction to phylum Tardigrada - Review Yashas R Devasurmutt1, Arpitha B M1* 1: R & D Centre, Department of Biotechnology, Dayananda Sagar College of Engineering, Bangalore, India 1*: Corresponding Author: Arpitha B M Abstract: Tardigrades popularly known as water bears are In cryptobiosis (extreme form of anabiosis), the metabolism is micrometazoans with four pairs of lobopod legs. They are the undetectable and the animal is known as tun in this phase. organisms which can live in extreme conditions and are known to Tuns have been known to survive very harsh environmental survive in vacuum and space without protection. Tardigardes conditions such as immersion in helium at -272° C (-458° F) survive in lichens and mosses, usually associated with water film or heating temperatures at 149° C (300° F), exposure to very on mosses, liverworts, and lichens. More species are found in high ionizing radiation and toxic chemical substances and milder environments such as meadows, ponds and lakes. They long durations without oxygen. [4] Figure 2 illustrates the are the first known species to survive in outer space. Tardigrades process of transition of the tardigrades[41]. are closely related to Arthropoda and nematodes based on their morphological and molecular analysis. The cryptobiosis of Figure 2: Transition process of Tardigrades Tardigrades have helped scientists to develop dry vaccines. They have been applied as research subjects in transplantology. Future research would help in more applications of tardigrades in the field of science. Keywords: Tardigrades, cryptobiosis, dry vaccines, Transplantology, space research I. INTRODUCTION ardigrade, a group of tiny arthropod-like animals having T four pairs of stubby legs with big claws, an oval stout body with a round back and lumbering gait. -

Tardigrade Hypsibius Klebelsbergimihelcic, 1959 (Tardigrada), Based on Material from the Btztal Alps, Austria
Hamburg, November 2003 I Mill. hamb. zool. Mu •. Inst. I Band 100 I S. 73-100 ISSN 0072 9612 Redescription and notes on the biology of the glacier tardigrade Hypsibius klebelsbergiMIHELcIC, 1959 (Tardigrada), based on material from the btztal Alps, Austria 1 2 3 HIERONYMUS DASTYCH , HANSJORG KRAUS & KONRAD THALER I UniversiUit Hamburg, Zoologisches Institut und Zoologisches Museum, Martin-Luther-King Platz 3, 20146 Hamburg, Germany; 2 Schloss-Tratzberg-StraBe 40, A-6200 Jenbach, Austria; 3 Institut fUr Zoologie & Limnologie, Universitilt Innsbruck, TechnikerstraBe 25, 6020 Innsbruck, Austria. ABSTRACT. - A redescription of a cryobiotic tardigrade, Hypsibli,J' /debe/sbergi MIHELCIC, 1959, is presented, based on material from cryoconite holes on the glacier Rotmoosfemer in the Otztal Alps (Austria). Much of the basic morphometric data of H. klebelsbergi is provided here for the first time and the bulk of available biological and ecological information about the species and its distribution is evaluated and discussed. The combination of some characters of H. klebelsbergi (e.g., the shape of anterior apophyses of the mouth tube and of the claws) indicates its separate generic status. A bisexual (amphimictic) reproduction mode for H. /debe/sbergi is implied. The latter and the taxonomic position of the species, including its possible synonymy with H. janelscheld Ramazzotti, 1968, known only from a Himalayan glacier, require further studies. KEYWORDS: Tardigrada, Hypsibills Idebelsbergl: redescription, SEM, taxonomy, glaciers, cryo conite holes, cryobionl, ecology, the Alps, Austria. Introduction Only a few invertebrate taxa dwell permanently on glaciers, where all available habitats are characterized by harsh environmental conditions. Cryoconite holes (= Kryokonitlocher, Mittagslocher), are aquatic microcaverns that occur on the ice surface (Fig. -

Will the Antarctic Tardigrade Acutuncus Antarcticus Be Able to Withstand
© 2018. Published by The Company of Biologists Ltd | Journal of Experimental Biology (2018) 221, jeb160622. doi:10.1242/jeb.160622 RESEARCH ARTICLE Will the Antarctic tardigrade Acutuncus antarcticus be able to withstand environmental stresses related to global climate change? Ilaria Giovannini1,*, Tiziana Altiero2,*, Roberto Guidetti1 and Lorena Rebecchi1,‡ ABSTRACT continental Antarctica is permanently covered by snow or ice, with Because conditions in continental Antarctica are highly selective and the only exception being ice-free terrestrial habitats restricted to extremely hostile to life, its biota is depauperate, but well adapted to coasts and inland nunataks. The snow covers terrestrial habitats for – live in this region. Global climate change has the potential to impact much of the year, and the frequency of daily freeze thaw events on continental Antarctic organisms because of increasing temperatures land is often unpredictable, sometimes occurring over hours, and ultraviolet radiation. This research evaluates how ongoing minutes or even more frequently (Convey, 1997; Wall, 2007; climate changes will affect Antarctic species, and whether Antarctic Convey et al., 2014). Moreover, the temperature is normally close to organisms will be able to adapt to the new environmental conditions. 0°C, with a narrow daily high temperature range, so temperatures Tardigrades represent one of the main terrestrial components suitable for life cycle activities are restricted to only a few weeks of Antarctic meiofauna; therefore, the pan-Antarctic tardigrade during the Antarctic summer (Convey et al., 2014; Everatt et al., Acutuncus antarcticus was used as model to predict the fate of 2014). As a consequence of these peculiar environmental Antarctic meiofauna threatened by climate change. -

Conference Program
WELCOME TO TARDIGRADA 2018 14TH INTERNATIONAL SYMPOSIUM ON TARDIGRADA CONFERENCE PROGRAM Symposi nal um tio o a n n Ta r r te d n i I g r h a t d 4 a 1 COPENHAGEN BIOCENTER, DENMARK www.tardigrada2018.org U N I V E R S I T Y O F C O P E N H A G E N FACULTY OF SCIENCE WELCOME 14th International Symposium on Tardigrada Welcome to Tardigrada 2018 International tardigrade symposia take place every three years and represent the greatest scientific forum on tardigrades. We are pleased to welcome you to Copenhagen and the 14th International Symposium on Tardigrada and it is with pleasure that we announce a new record in the number of participants with 28 countries represented at Tardigrada 2018. During the meeting 131 abstracts will be presented. The electronic abstract book is available for download from the Symposium website - www.tardigrada2018.org - and will be given to conference attendees on a USB stick during registration. Organising Committee 14th International Tardigrade Symposium, Copenhagen 2018 Chair Nadja Møbjerg (University of Copenhagen, Denmark) Local Committee Hans Ramløv (Roskilde University, Denmark), Jesper Guldberg Hansen (University of Copenhagen, Denmark), Jette Eibye-Jacobsen (University of Copenhagen, Denmark/ Birkerød Gymnasium), Lykke Keldsted Bøgsted Hvidepil (University of Copenhagen, Denmark), Maria Kamilari (University of Copenhagen, Denmark), Reinhardt Møbjerg Kristensen (University of Copenhagen, Denmark), Thomas L. Sørensen-Hygum (University of Copenhagen, Denmark) International Committee Ingemar Jönsson (Kristianstad University, Sweden), Łukasz Kaczmarek (A. Mickiewicz University, Poland) Łukasz Michalczyk (Jagiellonian University, Poland), Lorena Rebecchi (University of Modena and Reggio Emilia, Italy), Ralph O. -

Sponges Are Highly Resistant to Radiation Exposure and Cancer
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.17.435910; this version posted March 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Sponges are highly resistant to radiation exposure and cancer Angelo Fortunato1,2,3†, Jake Taylor1,2,3, Jonathan Scirone1,2,3, Athena Aktipis1,4* and Carlo C. Maley1,2,3* 1. Arizona Cancer Evolution Center, Arizona State University, 1001 S. McAllister Ave., Tempe, AZ, 85287, USA. 2. Biodesign Center for Biocomputing, Security and Society, Arizona State University, 727 E. Tyler St.,Tempe, AZ 85281, USA. 3. School of Life Sciences, Arizona State University, 427 East Tyler Mall, Tempe, AZ 85287, USA. 4. Department of Psychology, Arizona State University, Tempe, AZ, USA. † Corresponding author * co-senior authors bioRxiv preprint doi: https://doi.org/10.1101/2021.03.17.435910; this version posted March 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract There are no reports of cancer in sponges, despite them having somatic cell turnover, long lifespans and no specialized adaptive immune cells. In order to investigate whether sponges are cancer resistant, we exposed a species of sponge, Tethya wilhelma, to X-rays. We found that T. wilhelma can withstand 600 Gy of X-ray radiation. That is approximately 100 times the lethal dose for humans. A single high dose of X-rays did not induce cancer in sponges, providing the first experimental evidence of cancer resistance in the phylum, Porifera. -

Volume 2, Chapter 4-7C: Invertebrates: Rotifer Taxa
Glime, J. M. 2017. Invertebrates: Rotifer Taxa – Monogononta. Chapt. 4-7c. In: Glime, J. M. Bryophyte Ecology. Volume 2. 4-7c-1 Bryological Interaction. Ebook sponsored by Michigan Technological University and the International Association of Bryologists. Last updated 18 July 2020 and available at <http://digitalcommons.mtu.edu/bryophyte-ecology2/>. CHAPTER 4-7c INVERTEBRATES: ROTIFER TAXA – MONOGONONTA TABLE OF CONTENTS Notommatidae ............................................................................................................................................ 4-7c-2 Cephalodella ....................................................................................................................................... 4-7c-2 Drilophaga ........................................................................................................................................ 4-7c-10 Enteroplea ......................................................................................................................................... 4-7c-11 Eosphora ........................................................................................................................................... 4-7c-11 Eothinia ............................................................................................................................................. 4-7c-12 Monommata ...................................................................................................................................... 4-7c-12 Notommata ....................................................................................................................................... -

"Phoronida". In: Encyclopedia of Life Science
Phoronida Introductory article Christian C Emig, Centre d’Oce´anologie de Marseille CNRS, Marseille, France Article Contents . Basic Design The Phoronida, divided into two genera and 10 species, is a small marine group, which . Diversity and Lifestyles belongs to the phylum Lophophorata. Fossil History and Phylogeny Basic Design complexity of the lophophore from an oval to a helicoidal, The Phoronida is an exclusively marine group with a sessile through a horseshoe and spiral shape. This is related to an vermiform body enclosed in a tube (Figure 1) The body is increase in the number of tentacles, which is proportional composed of three distinct parts (prosome, mesosome and to the general body size. metasome), each containing its own coelomic cavity. The The metasome (or trunk) is slender and cylindrical, with prosome forms the epistome, a fold overhanging the mouth a bulb-like posterior end (or ampulla) that anchors the dorsally. The mesosome bears the lophophore, with the body in the rear end of the tube. It is separated from the rest mouth lying between its two rows of tentacles. The of the body by the diaphragm, a thick transverse septum lophophore is a terminal, bilaterally symmetrical, tentacle located behind the lophophore. crown, each tentacle having complex arrays of cilia for The digestive tract is U-shaped, bringing the anus close filter-feeding. Lophophore shape is a fairly constant to the mouth. The descending branch is divided into a short feature within each species, and there is an increasing oesophagus, followed by a long prestomach, then a stomach surrounded by a blood plexus. -
S I Section 4
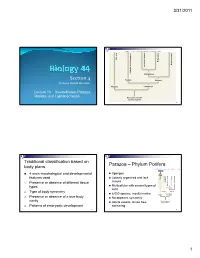
3/31/2011 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Porifera Ecdysozoa Deuterostomia Lophotrochozoa Cnidaria and Ctenophora Cnidaria and Protostomia SSiection 4 Radiata Bilateria Professor Donald McFarlane Parazoa Eumetazoa Lecture 13 Invertebrates: Parazoa, Radiata, and Lophotrochozoa Ancestral colonial choanoflagellate 2 Traditional classification based on Parazoa – Phylum Porifera body plans Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Parazoa 4 main morphological and developmental Sponges features used Loosely organized and lack Porifera Ecdysozoa Cnidaria and Ctenophora tissues euterostomia photrochozoa D 1. o Presence or absence of different tissue L Multicellular with several types of types Protostomia cells 2. Type of body symmetry Radiata Bilateria 8,000 species, mostly marine Parazoa Eumetazoa 3. Presence or absence of a true body No apparent symmetry cavity Ancestral colonial Adults sessile, larvae free- choanoflagellate 4. Patterns of embryonic development swimming 3 4 1 3/31/2011 Water drawn through pores (ostia) into spongocoel Flows out through osculum Reproduce Choanocytes line spongocoel Sexually Most hermapppgggphrodites producing eggs and sperm Trap and eat small particles and plankton Gametes are derived from amoebocytes or Mesohyl between choanocytes and choanocytes epithelial cells Asexually Amoebocytes absorb food from choanocytes, Small fragment or bud may detach and form a new digest it, and carry -

Tardigrades Colonise Antarctica?
This electronic thesis or dissertation has been downloaded from Explore Bristol Research, http://research-information.bristol.ac.uk Author: Short, Katherine A Title: Life in the extreme when did tardigrades colonise Antarctica? General rights Access to the thesis is subject to the Creative Commons Attribution - NonCommercial-No Derivatives 4.0 International Public License. A copy of this may be found at https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode This license sets out your rights and the restrictions that apply to your access to the thesis so it is important you read this before proceeding. Take down policy Some pages of this thesis may have been removed for copyright restrictions prior to having it been deposited in Explore Bristol Research. However, if you have discovered material within the thesis that you consider to be unlawful e.g. breaches of copyright (either yours or that of a third party) or any other law, including but not limited to those relating to patent, trademark, confidentiality, data protection, obscenity, defamation, libel, then please contact [email protected] and include the following information in your message: •Your contact details •Bibliographic details for the item, including a URL •An outline nature of the complaint Your claim will be investigated and, where appropriate, the item in question will be removed from public view as soon as possible. 1 Life in the Extreme: when did 2 Tardigrades Colonise Antarctica? 3 4 5 6 7 8 9 Katherine Short 10 11 12 13 14 15 A dissertation submitted to the University of Bristol in accordance with the 16 requirements for award of the degree of Geology in the Faculty of Earth 17 Sciences, September 2020. -

Antarctic Bdelloid Rotifers: Diversity, Endemism and Evolution
1 Antarctic bdelloid rotifers: diversity, endemism and evolution 2 3 Introduction 4 5 Antarctica’s ecosystems are characterized by the challenges of extreme environmental 6 stresses, including low temperatures, desiccation and high levels of solar radiation, all of 7 which have led to the evolution and expression of well-developed stress tolerance features in 8 the native terrestrial biota (Convey, 1996; Peck et al., 2006). The availability of liquid water, 9 and its predictability, is considered to be the most important driver of biological and 10 biodiversity processes in the terrestrial environments of Antarctica (Block et al., 2009; 11 Convey et al., 2014). Antarctica’s extreme conditions and isolation combined with the over- 12 running of many, but importantly not all, terrestrial and freshwater habitats by ice during 13 glacial cycles, underlie the low overall levels of diversity that characterize the contemporary 14 faunal, floral and microbial communities of the continent (Convey, 2013). Nevertheless, in 15 recent years it has become increasingly clear that these communities contain many, if not a 16 majority, of species that have survived multiple glacial cycles over many millions of years 17 and undergone evolutionary radiation on the continent itself rather than recolonizing from 18 extra-continental refugia (Convey & Stevens, 2007; Convey et al., 2008; Fraser et al., 2014). 19 With this background, high levels of endemism characterize the majority of groups that 20 dominate the Antarctic terrestrial fauna, including in particular Acari, Collembola, Nematoda 21 and Tardigrada (Pugh & Convey, 2008; Convey et al., 2012). 22 The continent of Antarctica is ice-bound, and surrounded and isolated from the other 23 Southern Hemisphere landmasses by the vastness of the Southern Ocean.