Analyses of Reticulate Evolution in the Apogamous Species of The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Seabirds in the Stomach Contents of Black Rats Rattus Rattus on Higashijima, the Ogasawara (Bonin) Islands, Japan
Yabe et al.: Seabirds in stomachs of Black Rats 293 SEABIRDS IN THE STOMACH CONTENTS OF BLACK RATS RATTUS RATTUS ON HIGASHIJIMA, THE OGASAWARA (BONIN) ISLANDS, JAPAN TATSUO YABE1,2, TAKUMA HASHIMOTO3, MASAAKI TAKIGUCHI3, MASANARI AOKI3 & KAZUTO KAWAKAMI4 1Rat Control Consulting, 1380–6 Fukuda, Yamato, Kanagawa, 242-0024, Japan ([email protected]) 2Tropical Rat Control Committee, c/o Overseas Agricultural Development Association, 8-10-32 Akasaka, Minato-Ku, Tokyo, 107-0052, Japan 3Japan Wildlife Research Center, 3-10-10 Shitaya, Taito-Ku, Tokyo, 110-8676, Japan 4Forestry and Forest Products Research Institute, 1 Matsunosato, Tsukuba, Ibaraki, 305-8687, Japan Received 11 September 2008, accepted 7 July 2009 Eradication programs for invasive rodents have accelerated since On Higashijima, Ogasawara Islands, rats are threatening the island pioneering efforts by New Zealand biologists in the 1970s to species. Seabirds such as Bulwer’s Petrels Bulweria bulwerii conserve not only seabird populations but also island ecosystems and Tristram’s Storm-Petrels are threatened by Black Rats. Ten (Howald et al. 2007, Rauzon 2007). In Japan, Black Rat Rattus carcasses of Bulwer’s Petrels were found for the first time on the rattus eradication programs have been carried out recently in the island in July 2005 (K. Horikoshi pers. comm.). Pictures taken Ogasawara (Bonin) Islands (Hashimoto 2009, Makino 2009). The during the period between June and October 2006 with automatic Ogasawara Islands are known to be a sanctuary for birds, including cameras (and the amount of rat tooth marks on bones) suggested several indigenous or endangered species such as Columba janthina that Black Rats preyed on more than 1000 Bulwer’s Petrels and their nitens, Apalopteron familiare, Tristram’s Storm-Petrel Oceanodroma eggs, and that they also ate Tristram’s Storm-Petrels (Horikoshi et tristrami, Matsudaira’s Storm-Petrel O. -

Vaitoskirjascientific MASCULINITY and NATIONAL IMAGES IN
Faculty of Arts University of Helsinki, Finland SCIENTIFIC MASCULINITY AND NATIONAL IMAGES IN JAPANESE SPECULATIVE CINEMA Leena Eerolainen DOCTORAL DISSERTATION To be presented for public discussion with the permission of the Faculty of Arts of the University of Helsinki, in Room 230, Aurora Building, on the 20th of August, 2020 at 14 o’clock. Helsinki 2020 Supervisors Henry Bacon, University of Helsinki, Finland Bart Gaens, University of Helsinki, Finland Pre-examiners Dolores Martinez, SOAS, University of London, UK Rikke Schubart, University of Southern Denmark, Denmark Opponent Dolores Martinez, SOAS, University of London, UK Custos Henry Bacon, University of Helsinki, Finland Copyright © 2020 Leena Eerolainen ISBN 978-951-51-6273-1 (paperback) ISBN 978-951-51-6274-8 (PDF) Helsinki: Unigrafia, 2020 The Faculty of Arts uses the Urkund system (plagiarism recognition) to examine all doctoral dissertations. ABSTRACT Science and technology have been paramount features of any modernized nation. In Japan they played an important role in the modernization and militarization of the nation, as well as its democratization and subsequent economic growth. Science and technology highlight the promises of a better tomorrow and future utopia, but their application can also present ethical issues. In fiction, they have historically played a significant role. Fictions of science continue to exert power via important multimedia platforms for considerations of the role of science and technology in our world. And, because of their importance for the development, ideologies and policies of any nation, these considerations can be correlated with the deliberation of the role of a nation in the world, including its internal and external images and imaginings. -

Perspectives of Research for Intangible Cultural Heritage
束 9mm Proceedings ISBN : 978-4-9909775-1-1 of the International Researchers Forum: Perspectives Research for Intangible Cultural Heritage towards a Sustainable Society Proceedings of International Researchers Forum: Perspectives of Research for Intangible Cultural Heritage towards a Sustainable Society 17-18 December 2019 Tokyo Japan Organised by International Research Centre for Intangible Cultural Heritage in the Asia-Pacific Region (IRCI), National Institutes for Cultural Heritage Agency for Cultural Affairs, Japan Co-organised by Tokyo National Research Institute for Cultural Properties, National Institutes for Cultural Heritage IRCI Proceedings of International Researchers Forum: Perspectives of Research for Intangible Cultural Heritage towards a Sustainable Society 17-18 December 2019 Tokyo Japan Organised by International Research Centre for Intangible Cultural Heritage in the Asia-Pacific Region (IRCI), National Institutes for Cultural Heritage Agency for Cultural Affairs, Japan Co-organised by Tokyo National Research Institute for Cultural Properties, National Institutes for Cultural Heritage Published by International Research Centre for Intangible Cultural Heritage in the Asia-Pacific Region (IRCI), National Institutes for Cultural Heritage 2 cho, Mozusekiun-cho, Sakai-ku, Sakai City, Osaka 590-0802, Japan Tel: +81 – 72 – 275 – 8050 Email: [email protected] Website: https://www.irci.jp © International Research Centre for Intangible Cultural Heritage in the Asia-Pacific Region (IRCI) Published on 10 March 2020 Preface The International Researchers Forum: Perspectives of Research for Intangible Cultural Heritage towards a Sustainable Society was organised by the International Research Centre for Intangible Cultural Heritage in the Asia-Pacific Region (IRCI) in cooperation with the Agency for Cultural Affairs of Japan and the Tokyo National Research Institute for Cultural Properties on 17–18 December 2019. -
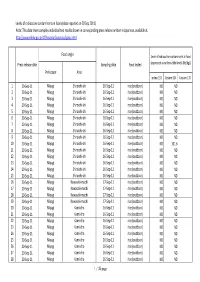
This Data Sheet Compiles Individual Test Results Shown in Corresponding
Levels of radioactive contaminants in foods (data reported on 20 Sep 2011) Note: This data sheet compiles individual test results shown in corresponding press release written in Japanese, available at http://www.mhlw.go.jp/stf/houdou/bukyoku/iyaku.html Food origin Level of radioactive contaminants in food Press release date Sampling date Food tested (expressed as radionuclide levels (Bq/kg)). Prefecture Area Iodine‐131 Cesium‐134 Cesium‐137 1 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 2 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 3 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 4 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 5 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 6 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 7 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 8 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 9 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 10 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND 101.6 11 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 12 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 13 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 14 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 15 20‐Sep‐11 Miyagi Shiroishi‐shi 16‐Sep‐11 rice (outdoor) ND ND 16 20‐Sep‐11 Miyagi Kawasaki‐machi 17‐Sep‐11 rice (outdoor) ND ND 17 20‐Sep‐11 Miyagi Kawasaki‐machi 17‐Sep‐11 rice (outdoor) ND ND 18 20‐Sep‐11 Miyagi -
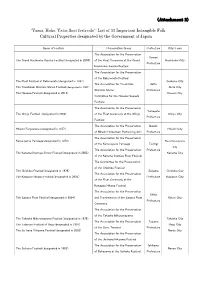
(Attachment 2)“Yama, Hoko, Yatai Float Festivals”: List of 33 Important
( ) Attachment 2 “Yama, Hoko, Yatai float festivals”: List of 33 Important Intangible Folk Cultural Properties designated by the Government of Japan Name of Festival Preservation Group Prefecture City/Town The Association for the Preservation Aomori The Grand Hachinohe Sansha Festival(designated in 2004) of the Float Ceremony at the Grand Hachinohe City Prefecture Hachinohe Sansha Festival The Association for the Preservation of the Kakunodate Festival The Float Festival of Kakunodate(designated in 1991) Senboku City The Association for Tsuchizaki Akita The Tsuchizaki Shimmei Shrine Festival(designated in 1997) Akita City Shimmei Shrine Prefecture The Hanawa Festival(designated in 2014) Kazuno City Committee for the Hanawa-bayashi Festival The Association for the Preservation Yamagata The Shinjo Festival (designated in 2009) of the Float Ceremony at the Shinjo Shinjo City Prefecture Festival The Association for the Preservation Ibaraki Hitachi Furyumono(designated in 1977) Hitachi City of Hitachi Hometown Performing Arts Prefecture The Association for the Preservation Karasuyama Yamaage(designated in 1979) Nasu Karasuyama of the Karasuyama Yamaage Tochigi City The Association for the Preservation Prefecture The Kanuma Imamiya Shrine Festival(designated in 2003) Kanuma City of the Kanuma Imamiya Float Festival The Committee for the Preservation of the Chichibu Festival The Chichibu Festival(designated in 1979) Saitama Chichibu City The Association for the Preservation The Kawagoe Hikawa Festival(designated in 2005) Prefecture Kawagoe City -

UNIVERSITY of HAWAII LIBRARY. NARRATIVES OF
,UNIVERSITY Of HAWAII LIBRARY. NARRATIVES OF SPACE AND PLACE IN THREE WORKS BY NAKAGAMI KENJI A THESIS SUBMITTED TO THE GRADUATE DIVISION OF THE UNIVERSITY OF HAWAI'I IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF ARTS IN EAST ASIAN LANGUAGES AND LITERATURES (JAPANESE) AUGUST 2005 By Joshua Petitto Thesis Committee: Nobuko Ochner, Chairperson Lucy Lower Arthur Thornhill © Copyright 2005 by Joshua Petitto 111 TABLE OF CONTENTS Chapter 1 - Introduction 1.1 Nakagami's Style , 1 1.2 Literature Review 3 1.3 Summary ofthe Chapters ,. '" 8 1.4 Misaki, "Garyl1san," and "Wara no ie"............................................. 9 Chapter 2 - Down the Ever-Winding Narrative Path 2.1 Introduction........................................................................... 11 2.2 The Problem ofNarrative 11 2.3 Taking on the "Pig ofNarrative". 17 2.5 Conclusion: Monogatari and Genealogy....................................... 20 Chapter 3 - The Ambivalence ofNakagami's Space 3.1 Introduction 3.1.1 Conceptualizing Space 23 3.1.2 The Space ofthe Roji 28 3.2 A History ofKumano 30 3.3 The Ambivalence ofNakagami's Space 3.3.1 Attempted Assertion over Space through Ritual.................. 35 3.3.2 The Dominance ofSpace............................................... 42 3.3.3 Gendered Space and Origin............................................ 44 3.3.4 Reconstructing Space................................................... 47 3.4 Conclusion: The Same Struggle by Another Name............................. 49 Chapter 4 - The Place ofMemory 4.1 Introduction 4.1.1 Space and Place 51 4.1.2 The Recovery ofOrigin..................................... .. 54 4.2 Narratives ofPlace in Misaki, "Garyl1san," and "Wara no ie" 4.2.1 The Brother and Father in Misaki 58 4.2.2 Garyl1 Mountain and Toshihisa 66 4.2.3 The Rediscovery ofDifference in "Wara no ie" 75 4.3 Conclusion: Place, Narrative, and Rememory 80 Chapter 5 - Coming Full Circle: Space, Narrative, and the Next Roji 5.1 Introduction 84 5.2 The Emperor System and Capital................................................ -

Illustration and the Visual Imagination in Modern Japanese Literature By
Eyes of the Heart: Illustration and the Visual Imagination in Modern Japanese Literature By Pedro Thiago Ramos Bassoe A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor in Philosophy in Japanese Literature in the Graduate Division of the University of California, Berkeley Committee in Charge: Professor Daniel O’Neill, Chair Professor Alan Tansman Professor Beate Fricke Summer 2018 © 2018 Pedro Thiago Ramos Bassoe All Rights Reserved Abstract Eyes of the Heart: Illustration and the Visual Imagination in Modern Japanese Literature by Pedro Thiago Ramos Bassoe Doctor of Philosophy in Japanese Literature University of California, Berkeley Professor Daniel O’Neill, Chair My dissertation investigates the role of images in shaping literary production in Japan from the 1880’s to the 1930’s as writers negotiated shifting relationships of text and image in the literary and visual arts. Throughout the Edo period (1603-1868), works of fiction were liberally illustrated with woodblock printed images, which, especially towards the mid-19th century, had become an essential component of most popular literature in Japan. With the opening of Japan’s borders in the Meiji period (1868-1912), writers who had grown up reading illustrated fiction were exposed to foreign works of literature that largely eschewed the use of illustration as a medium for storytelling, in turn leading them to reevaluate the role of image in their own literary tradition. As authors endeavored to produce a purely text-based form of fiction, modeled in part on the European novel, they began to reject the inclusion of images in their own work. -

Summary of Family Membership and Gender by Club MBR0018 As of November, 2008
Summary of Family Membership and Gender by Club MBR0018 as of November, 2008 Club Fam. Unit Fam. Unit Club Ttl. Club Ttl. District Number Club Name HH's 1/2 Dues Females Male TOTAL District 334 C 24664 ARAIMACHI L C 0 0 0 27 27 District 334 C 24665 ATAMI 0 0 0 22 22 District 334 C 24671 FUJI GAKUNAN 3 3 0 65 65 District 334 C 24672 FUJIEDA 0 0 0 45 45 District 334 C 24673 FUJINOMIYA 0 0 3 68 71 District 334 C 24674 FUKUROI 0 0 1 59 60 District 334 C 24676 GOTENBA 0 0 0 97 97 District 334 C 24677 HAINAN 0 0 1 49 50 District 334 C 24678 HAMAMATSU 0 0 3 132 135 District 334 C 24679 HAMAKITA 0 0 1 40 41 District 334 C 24682 HIGASHIIZU 0 0 1 21 22 District 334 C 24683 OKUHAMANAKO L C 0 0 0 20 20 District 334 C 24693 ITO 0 0 0 42 42 District 334 C 24694 IWATA 0 0 4 65 69 District 334 C 24695 KAKEGAWA 0 0 1 45 46 District 334 C 24697 KANBARA 0 0 0 44 44 District 334 C 24702 KAWANE 0 0 0 53 53 District 334 C 24708 KOSAI 0 0 2 53 55 District 334 C 24720 MISHIMA 0 0 0 34 34 District 334 C 24722 MORIMACHI L C 0 0 3 41 44 District 334 C 24723 NAGAIZUMI 0 0 0 11 11 District 334 C 24747 NISHIIZU 0 0 0 19 19 District 334 C 24749 NUMAZU 0 0 0 70 70 District 334 C 24750 NUMAZU SENBON 0 0 0 43 43 District 334 C 24753 OHITO 0 0 0 28 28 District 334 C 24761 OYAMA 0 0 0 27 27 District 334 C 24765 SHIMIZUCHO 0 0 0 15 15 District 334 C 24766 SHIBAKAWA 0 0 0 19 19 District 334 C 24767 SHIZUOKA TACHIBANA 0 0 0 52 52 District 334 C 24768 SHUZENJI 0 0 0 30 30 District 334 C 24770 SHIMADA 0 0 2 71 73 District 334 C 24771 SHIMIZU HAGOROMO 0 0 0 33 33 District 334 -

Ogasawara) Archipelago, Japan, and Its Identification Using Molecular Sequences from a Herbarium Specimen Collected More Than 100 Years Ago
ISSN 1346-7565 Acta Phytotax. Geobot. 70 (3): 149–158 (2019) doi: 10.18942/apg.201901 Rediscovery of Liparis hostifolia (Orchidaceae) on Minami-iwo-to Island in the Bonin (Ogasawara) Archipelago, Japan, and its Identification Using Molecular Sequences from a Herbarium Specimen Collected more than 100 Years Ago 1,† 2,† 3 4 Koji TaKayama , Chie TsuTsumi , Dairo KawaguChi , hiDeToshi KaTo anD 2,* Tomohisa yuKawa 1Department of Botany, Graduate School of Science, Kyoto University, Kitashirakawa Oiwake-cho, Sakyo-ku, Kyoto 606-8502, Japan; 2 Department of Botany, National Museum of Nature and Science, 4-1-1 Amakubo, Tsukuba 305- 0005, Japan. *[email protected] (author for correspondence); 3 Ogasawara Islands Branch Office, Tokyo Metropolitan Government, Nishimachi, Chichi-jima, Ogasawara-mura, Tokyo 100-2101, Japan; 4 Department of Biological Sciences, Tokyo Metropolitan University, 1-1 Minami Osawa, Hachioji, Tokyo 192-0397, Japan. † These authors contributed equally to this work Liparis hostifolia (Orchidaceae) on Minami-iwo-to Island in the Bonin (Ogasawara) Archipelago was rediscovered for the first time in 79 years during a field survey in 2017. Its identity was confirmed by morphological comparison and DNA extractions from herbarium specimens collected between 1914 and 1938. Results from the molecular phylogenetic analyses demonstrated that L. hostifolia belongs to the L. makinoana complex. In comparison with other members of the L. makinoana complex, the broadly ovate labellum, short dormancy period, and flowering from November to March are unique characteristics of L. hostifolia. Results from the molecular phylogenetic analyses also suggested that L. hostifolia has had a long-isolated history in the Bonin Archipelago and probably migrated from temperate East Asia. -

Forest Structures, Composition, and Distribution on a Pacific Island, with Reference to Ecological Release and Speciation!
Pacific Science (1991), vol. 45, no. 1: 28-49 © 1991 by University of Hawaii Press. All rights reserved Forest Structures, Composition, and Distribution on a Pacific Island, with Reference to Ecological Release and Speciation! YOSHIKAZU SHIMIZU2 AND HIDEO TABATA 3 ABSTRACT: Native forest and scrub of Chichijima, the largest island in the Bonins, were classified into five types based on structural features: Elaeocarpus Ardisia mesic forest, 13-16 m high, dominated by Elaeocarpus photiniaefolius and Ardisia sieboldii; Pinus-Schima mesic forest, 12-16 m high, consisting of Schima mertensiana and an introduced pine , Pinus lutchuensis; Rhaphiolepis Livistonia dry forest, 2-6 m high, mainly occupied by Rhaphiolepis indica v. integerrima; Distylium-Schima dry forest, 3-8 m high, dominated by Distylium lepidotum and Schima mertensiana; and Distylium-Pouteria dry scrub, 0.3 1.5 m high, mainly composed of Distylium lepidotum. A vegetation map based on this classification was developed. Species composition and structural features of each type were analyzed in terms of habitat condition and mechanisms of regeneration. A group of species such as Pouteria obovata, Syzgygium buxifo lium, Hibiscus glaber, Rhaphiolepis indica v. integerrima, and Pandanus boninen sis, all with different growth forms from large trees to stunted shrubs, was subdominant in all vegetation types. Schima mertensiana , an endemic pioneer tree, occurred in both secondary forests and climax forests as a dominant canopy species and may be an indication of "ecological release," a characteristic of oceanic islands with poor floras and little competitive pressure. Some taxonomic groups (Callicarpa, Symplocos, Pittosporum, etc.) have speciated in the under story of Distylium-Schima dry forest and Distylium-Pouteria dry scrub. -

A POPULAR DICTIONARY of Shinto
A POPULAR DICTIONARY OF Shinto A POPULAR DICTIONARY OF Shinto BRIAN BOCKING Curzon First published by Curzon Press 15 The Quadrant, Richmond Surrey, TW9 1BP This edition published in the Taylor & Francis e-Library, 2005. “To purchase your own copy of this or any of Taylor & Francis or Routledge’s collection of thousands of eBooks please go to http://www.ebookstore.tandf.co.uk/.” Copyright © 1995 by Brian Bocking Revised edition 1997 Cover photograph by Sharon Hoogstraten Cover design by Kim Bartko All rights reserved. No part of this book may be reproduced, stored in a retrieval system, or transmitted in any form or by any means, electronic, mechanical, photocopying, recording, or otherwise, without the prior permission of the publisher. British Library Cataloguing in Publication Data A catalogue record for this book is available from the British Library ISBN 0-203-98627-X Master e-book ISBN ISBN 0-7007-1051-5 (Print Edition) To Shelagh INTRODUCTION How to use this dictionary A Popular Dictionary of Shintō lists in alphabetical order more than a thousand terms relating to Shintō. Almost all are Japanese terms. The dictionary can be used in the ordinary way if the Shintō term you want to look up is already in Japanese (e.g. kami rather than ‘deity’) and has a main entry in the dictionary. If, as is very likely, the concept or word you want is in English such as ‘pollution’, ‘children’, ‘shrine’, etc., or perhaps a place-name like ‘Kyōto’ or ‘Akita’ which does not have a main entry, then consult the comprehensive Thematic Index of English and Japanese terms at the end of the Dictionary first. -

What Is Dewa Sanzan? the Spiritual Awe-Inspiring Mountains in the Tohoku Area, Embracing Peopleʼs Prayers… from the Heian Period, Mt.Gassan, Mt.Yudono and Mt
The ancient road of Dewa Rokujurigoegoe Kaido Visit the 1200 year old ancient route! Sea of Japan Yamagata Prefecture Tsuruoka City Rokujurigoe Kaido Nishikawa Town Asahi Tourism Bureau 60-ri Goe Kaido Tsuruoka City, Yamagata Prefecture The Ancient Road “Rokujuri-goe Kaido” Over 1200 years, this road has preserved traces of historical events “Rokujuri-goe Kaido,” an ancient road connecting the Shonai plain and the inland area is said to have opened about 1200 years ago. This road was the only road between Shonai and the inland area. It was a precipitous mountain road from Tsuruoka city to Yamagata city passing over Matsune, Juo-toge, Oami, Sainokami-toge, Tamugimata and Oguki-toge, then going through Shizu, Hondoji and Sagae. It is said to have existed already in ancient times, but it is not clear when this road was opened. The oldest theory says that this road was opened as a governmental road connecting the Dewa Kokufu government which was located in Fujishima town (now Tsuruoka city) and the county offices of the Mogami and Okitama areas. But there are many other theories as well. In the Muromachi and Edo periods, which were a time of prosperity for mountain worship, it became a lively road with pilgrims not only from the local area,but also from the Tohoku Part of a list of famous places in Shonai second district during the latter half of the Edo period. and Kanto areas heading to Mt. Yudono as “Oyama mairi” (mountain pilgrimage) custom was (Stored at the Native district museum of Tsuruoka city) booming.