Poised Lineage Specification in Multipotential Hematopoietic Stem
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

CDH12 Cadherin 12, Type 2 N-Cadherin 2 RPL5 Ribosomal
5 6 6 5 . 4 2 1 1 1 2 4 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 A A A A A A A A A A A A A A A A A A A A C C C C C C C C C C C C C C C C C C C C R R R R R R R R R R R R R R R R R R R R B , B B B B B B B B B B B B B B B B B B B , 9 , , , , 4 , , 3 0 , , , , , , , , 6 2 , , 5 , 0 8 6 4 , 7 5 7 0 2 8 9 1 3 3 3 1 1 7 5 0 4 1 4 0 7 1 0 2 0 6 7 8 0 2 5 7 8 0 3 8 5 4 9 0 1 0 8 8 3 5 6 7 4 7 9 5 2 1 1 8 2 2 1 7 9 6 2 1 7 1 1 0 4 5 3 5 8 9 1 0 0 4 2 5 0 8 1 4 1 6 9 0 0 6 3 6 9 1 0 9 0 3 8 1 3 5 6 3 6 0 4 2 6 1 0 1 2 1 9 9 7 9 5 7 1 5 8 9 8 8 2 1 9 9 1 1 1 9 6 9 8 9 7 8 4 5 8 8 6 4 8 1 1 2 8 6 2 7 9 8 3 5 4 3 2 1 7 9 5 3 1 3 2 1 2 9 5 1 1 1 1 1 1 5 9 5 3 2 6 3 4 1 3 1 1 4 1 4 1 7 1 3 4 3 2 7 6 4 2 7 2 1 2 1 5 1 6 3 5 6 1 3 6 4 7 1 6 5 1 1 4 1 6 1 7 6 4 7 e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m -

A Promising Marker of Hormone Refractory Metastatic Prostate Cancer
Vol. 11, 2237–2243, March 15, 2005 Clinical Cancer Research 2237 Reg IV: A Promising Marker of Hormone Refractory Metastatic Prostate Cancer Zhennan Gu,2 Mark A. Rubin,4 Yu Yang,4 growing tumors that may not impact an individual’s natural life Samuel E. Deprimo,5 Hongjuan Zhao,5 span, although others are struck by rapidly progressive, metastatic tumors. Prostate-specific antigen screening is limited by a lack of Steven Horvath,1 James D. Brooks,5 4 2,3 specificity and an inability to predict which patients are at risk to Massimo Loda, and Robert E. Reiter develop hormone refractory metastatic disease. Recent studies Departments of 1Statistics and 2Urology, and the 3Molecular Biology advocating a lower prostate-specific antigen threshold for Institute, Geffen School of Medicine at University of California at diagnosis may increase the number of prostate cancer diagnoses Los Angeles, Los Angeles, California; 4Department of Pathology, Dana-Farber Cancer Institute, Harvard School of Medicine, Boston, and further complicate the identification of patients with indolent Massachusetts; and 5Department of Urology, Stanford University versus aggressive cancers (1). New serum and tissue markers that School of Medicine, Stanford, California correlate with clinical outcome or identify patients with potentially aggressive disease are urgently needed (2). Recent expression profiling studies suggest that expression ABSTRACT signatures for metastatic versus nonmetastatic tumors may reside The diagnosis and management of prostate cancer is in the primary tumor (2–4). Additional features that predispose hampered by the absence of markers capable of identifying tumors to metastasize to specific organs may also be present at patients with metastatic disease. -

Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association Between BMI and Adult-Onset Non- Atopic
Supplementary Material DNA Methylation in Inflammatory Pathways Modifies the Association between BMI and Adult-Onset Non- Atopic Asthma Ayoung Jeong 1,2, Medea Imboden 1,2, Akram Ghantous 3, Alexei Novoloaca 3, Anne-Elie Carsin 4,5,6, Manolis Kogevinas 4,5,6, Christian Schindler 1,2, Gianfranco Lovison 7, Zdenko Herceg 3, Cyrille Cuenin 3, Roel Vermeulen 8, Deborah Jarvis 9, André F. S. Amaral 9, Florian Kronenberg 10, Paolo Vineis 11,12 and Nicole Probst-Hensch 1,2,* 1 Swiss Tropical and Public Health Institute, 4051 Basel, Switzerland; [email protected] (A.J.); [email protected] (M.I.); [email protected] (C.S.) 2 Department of Public Health, University of Basel, 4001 Basel, Switzerland 3 International Agency for Research on Cancer, 69372 Lyon, France; [email protected] (A.G.); [email protected] (A.N.); [email protected] (Z.H.); [email protected] (C.C.) 4 ISGlobal, Barcelona Institute for Global Health, 08003 Barcelona, Spain; [email protected] (A.-E.C.); [email protected] (M.K.) 5 Universitat Pompeu Fabra (UPF), 08002 Barcelona, Spain 6 CIBER Epidemiología y Salud Pública (CIBERESP), 08005 Barcelona, Spain 7 Department of Economics, Business and Statistics, University of Palermo, 90128 Palermo, Italy; [email protected] 8 Environmental Epidemiology Division, Utrecht University, Institute for Risk Assessment Sciences, 3584CM Utrecht, Netherlands; [email protected] 9 Population Health and Occupational Disease, National Heart and Lung Institute, Imperial College, SW3 6LR London, UK; [email protected] (D.J.); [email protected] (A.F.S.A.) 10 Division of Genetic Epidemiology, Medical University of Innsbruck, 6020 Innsbruck, Austria; [email protected] 11 MRC-PHE Centre for Environment and Health, School of Public Health, Imperial College London, W2 1PG London, UK; [email protected] 12 Italian Institute for Genomic Medicine (IIGM), 10126 Turin, Italy * Correspondence: [email protected]; Tel.: +41-61-284-8378 Int. -

IGLL1 Polyclonal Antibody
PRODUCT DATA SHEET Bioworld Technology,Inc. IGLL1 polyclonal antibody Catalog: BS65198 Host: Rabbit Reactivity: Human BackGround: Storage&Stability: immunoglobulin lambda like polypeptide 1(IGLL1) Ho- Store at 4°C short term. Aliquot and store at -20°C long mo sapiens The preB cell receptor is found on the surface term. Avoid freeze-thaw cycles. of proB and preB cells, where it is involved in transduc- Specificity: tion of signals for cellular proliferation, differentiation Lambda 5 Polyclonal Antibody detects endogenous levels from the proB cell to the preB cell stage, allelic exclusion of Lambda 5 protein. at the Ig heavy chain gene locus, and promotion of Ig DATA: light chain gene rearrangements. The preB cell receptor is composed of a membrane-bound Ig mu heavy chain in association with a heterodimeric surrogate light chain. This gene encodes one of the surrogate light chain subu- nits and is a member of the immunoglobulin gene super- family. This gene does not undergo rearrangement. Muta- tions in this gene can result in B cell deficiency and agammaglobulinemia, an autosomal recessive disease in Western Blot analysis of KB cells using Lambda 5 Polyclonal Anti- which few or no gamma globulins or antibodies are made. body.. Secondary antibody(catalog#:RS0002) was diluted at 1:20000 Two transcript variants encoding different isoforms have been found for this gene. [provided by RefSeq, Jul 2008], Product: Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide. Molecular Weight: ~ 23 kDa Swiss-Prot: Immunohistochemical analysis of paraffin-embedded human-lymph, P15814 antibody was diluted at 1:100 Purification&Purity: Note: The antibody was affinity-purified from rabbit antiserum For research use only, not for use in diagnostic procedure. -

The Effect of Expression of Estrus at Artificial Insemination on Target Genes in the Endometrium, Conceptus and Corpus Luteum of Beef Cows
THE EFFECT OF EXPRESSION OF ESTRUS AT ARTIFICIAL INSEMINATION ON TARGET GENES IN THE ENDOMETRIUM, CONCEPTUS AND CORPUS LUTEUM OF BEEF COWS by Saeideh Davoodi A THESIS SUBMITTED IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF SCIENCE in The Faculty of Graduate and Postdoctoral Studies (Applied Animal Biology) THE UNIVERSITY OF BRITISH COLUMBIA (Vancouver) April 2015 © Saeideh Davoodi, 2015 Abstract The aim of this study was to test the effect of expression of estrus at artificial insemination (AI) on the endometrium, conceptus and corpus luteum (CL) gene expression. Twenty-three multiparous non-lactating Nelore cows were enrolled on an estradiol (E2) and progesterone (P4) based timed-AI protocol (AI = d 0), then slaughtered for endometrium, CL and conceptus collection on d 19. Body condition score (BCS), blood samples and ultrasound examination was performed on d 0, 7 and 18 of the experiment followed by RNA extraction and quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of 58 target genes. Data was checked for normality and analysed by ANOVA for repeated measures using proc GLM, MIXED and UNIVARIATE. Estrous expression had no correlation with parameters such as BCS, pre-ovulatory follicle and CL diameter, P4 concentration in plasma on d 7 and 18 after AI and IFN-tau concentration in the uterine flushing (P > 0.05); however, a significant increase was observed in conceptus size (P = 0.02; 38.3 ± 2.8 vs 28.2 ± 2.9). The majority of transcripts affected by estrous expression in the endometrium belong to the immune system and adhesion molecule family (MX1, MX2, MYL12A, MMP19, CXCL10, IGLL1 and SLPI) (P ≤ 0.05). -

Genetic Defects in B-Cell Development and Their Clinical Consequences H Abolhassani,1,2 N Parvaneh,1 N Rezaei,1 L Hammarström,2 a Aghamohammadi1
REVIEWS Genetic Defects in B-Cell Development and Their Clinical Consequences H Abolhassani,1,2 N Parvaneh,1 N Rezaei,1 L Hammarström,2 A Aghamohammadi1 1Research Center for Immunodeficiencies, Pediatrics Center of Excellence, Children’s Medical Center, Tehran University of Medical Sciences, Tehran, Iran 2Division of Clinical Immunology, Department of Laboratory Medicine, Karolinska Institutet at Karolinska University Hospital Huddinge, Stockholm, Sweden n Abstract Expression of selected genes in hematopoietic stem cells has been identified as a regulator of differentiation of B cells in the liver and bone marrow. Moreover, naïve B cells expressing surface immunoglobulin need other types of genes for antigen-dependent development in secondary lymphoid organs. Many advanced molecular mechanisms underlying primary antibody deficiencies in humans have been described. We provide an overview of the mutations in genes known to be involved in B-cell development and their clinical consequences. Key words: Genetic disorder. B-cell development. Primary antibody deficiencies. Clinical phenotypes. n Resumen Se ha identificado la expresión de genes seleccionados en las células pluripotenciales de médula ósea como reguladores de la diferenciación de las células B en el hígado y en médula ósea. Sin embargo, las células B naïve que expresan inmunoglubulinas de superficie, necesitan otros tipos de genes para su desarrollo en los órganos linfoides secundarios dependienteS de antígeno. Se han descrito muchos mecanismos moleculares avanzados que subrayan las inmunodeficiencias en humanos y esta revisión constituye una visión general de la mutación en todos los genes conocidos involucrados en el desarrollo de las células B y sus consecuencias clínicas. Palabras clave: Alteraciones genéticas. Desarrollo de las células B. -

Urinary Proteomics for the Early Diagnosis of Diabetic Nephropathy in Taiwanese Patients Authors
Urinary Proteomics for the Early Diagnosis of Diabetic Nephropathy in Taiwanese Patients Authors: Wen-Ling Liao1,2, Chiz-Tzung Chang3,4, Ching-Chu Chen5,6, Wen-Jane Lee7,8, Shih-Yi Lin3,4, Hsin-Yi Liao9, Chia-Ming Wu10, Ya-Wen Chang10, Chao-Jung Chen1,9,+,*, Fuu-Jen Tsai6,10,11,+,* 1 Graduate Institute of Integrated Medicine, China Medical University, Taichung, 404, Taiwan 2 Center for Personalized Medicine, China Medical University Hospital, Taichung, 404, Taiwan 3 Division of Nephrology and Kidney Institute, Department of Internal Medicine, China Medical University Hospital, Taichung, 404, Taiwan 4 Institute of Clinical Medical Science, China Medical University College of Medicine, Taichung, 404, Taiwan 5 Division of Endocrinology and Metabolism, Department of Medicine, China Medical University Hospital, Taichung, 404, Taiwan 6 School of Chinese Medicine, China Medical University, Taichung, 404, Taiwan 7 Department of Medical Research, Taichung Veterans General Hospital, Taichung, 404, Taiwan 8 Department of Social Work, Tunghai University, Taichung, 404, Taiwan 9 Proteomics Core Laboratory, Department of Medical Research, China Medical University Hospital, Taichung, 404, Taiwan 10 Human Genetic Center, Department of Medical Research, China Medical University Hospital, China Medical University, Taichung, 404, Taiwan 11 Department of Health and Nutrition Biotechnology, Asia University, Taichung, 404, Taiwan + Fuu-Jen Tsai and Chao-Jung Chen contributed equally to this work. Correspondence: Fuu-Jen Tsai, MD, PhD and Chao-Jung Chen, PhD FJ Tsai: Genetic Center, China Medical University Hospital, No.2 Yuh-Der Road, 404 Taichung, Taiwan; Telephone: 886-4-22062121 Ext. 2041; Fax: 886-4-22033295; E-mail: [email protected] CJ Chen: Graduate Institute of Integrated Medicine, China Medical University, No.91, Hsueh-Shih Road, 404, Taichung, Taiwan; Telephone: 886-4-22053366 Ext. -
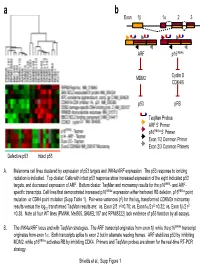
Taqman Probes ARF 5' Primer P16ink4a 5' Primer Exon 1/2
a b Exon 1β 1α 23 ARF p16INK4a MDM2 Cyclin D CDK4/6 p53 pRB TaqMan Probes ARF 5’ Primer p16INK4a 5’ Primer Exon 1/2 Common Primer Exon 2/3 Common Primers Defective p53 Intact p53 A. Melanoma cell lines clustered by expression of p53 targets and INK4a/ARF expression. The p53 response to ionizing radiation is indicated. Top cluster: Cells with intact p53 response show increased expression of the eight indicated p53 targets, and decreased expression of ARF. Bottom cluster: TaqMan and microarray results for the p16INK4- and ARF- specific transcripts. Cell lines that demonstrated increased p16INK4a expression either harbored RB deletion, p16INK4a point 2 mutation or CDK4 point mutation (Supp Table 1). Pair-wise variances (r ) for the log2 transformed CDKN2a microarray 2 2 2 results versus the log10 transformed TaqMan results are: vs. Exon 2/3 r =0.78; vs. Exon1α/2 r =0.32; vs. Exon 1β/2 r =0.38. Note all four WT lines (PMWK, Mel505, SKMEL187 and RPMI8322) lack evidence of p53 function by all assays. B. The INK4a/ARF locus and with TaqMan strategies. The ARF transcript originates from exon 1β while the p16INK4a transcript originates from exon 1α. Both transcripts splice to exon 2 but in alternate reading frames. ARF stabilizes p53 by inhibiting MDM2, while p16INK4a activates RB by inhibiting CDK4. Primers and TaqMan probes are shown for the real-time RT-PCR strategy. Shields et al., Supp Figure 1 SKMEL 28 U01 24h SKMEL WM2664 U01 48h WM2664 U01 24h 24 U01 48h SKMEL 24 U01 24h SKMEL 24 Untreated SKMEL 24 DMSO 48h SKMEL 24 DMSO 24h SKMEL WM2664 DMSO 24h WM2664 DMSO 48h WM2644 Untreated 28 Untreated SKMEL 28 DMSO 48h SKMEL 28 DMSO 24h SKMEL -3.00 -2.00 -1.00 0.00 1.00 2.00 3.00 relative to median expression Genes decreased by UO1 (863) HIF1A Hypoxia-inducible factor 1, alpha subunit NM_001530 RBBP8 Retinoblastoma binding protein 8 NM_002894 Homo sapiens, clone IMAGE:4337652, mRNA BC018676 EIF4EBP1 Eukaryotic translation initiation factor 4E binding protein EXOSC8 Exosome component 8 NM_181503 ENST00000321524 MCM7 MCM7 minichromosome maintenance deficient 7 (S. -

Anti-IGLL1 / Lambda 5 Antibody (ARG42961)
Product datasheet [email protected] ARG42961 Package: 50 μg anti-IGLL1 / Lambda 5 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes IGLL1 / Lambda 5 Tested Reactivity Hu, Ms, Rat Tested Application IHC-P, WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name IGLL1 / Lambda 5 Antigen Species Human Immunogen Recombinant protein corresponding to L38-S213 of Human IGLL1 / Lambda 5. Conjugation Un-conjugated Alternate Names IGLL; Immunoglobulin omega polypeptide; 14.1; IGVPB; Immunoglobulin lambda-like polypeptide 1; Immunoglobulin-related protein 14.1; CD179 antigen-like family member B; IGLJ14.1; Ig lambda-5; CD antigen CD179b; AGM2; CD179b; IGO; IGL5; IGL1; VPREB2 Application Instructions Application table Application Dilution IHC-P 1:200 - 1:1000 WB 1:500 - 1:2000 Application Note IHC-P: Antigen Retrieval: Heat mediation was performed in Citrate buffer (pH 6.0) for 20 min. * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Calculated Mw 23 kDa Observed Size ~ 26 kDa Properties Form Liquid Purification Affinity purification with immunogen. Buffer 0.2% Na2HPO4, 0.9% NaCl, 0.05% Sodium azide and 4% Trehalose. Preservative 0.05% Sodium azide Stabilizer 4% Trehalose Concentration 0.5 - 1 mg/ml Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated www.arigobio.com 1/3 freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use.