Characterizing Industrial and Artisanal Fishing Vessel Catch Composition Using Environmental DNA and Satellite-Based Tracking Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Skeletal Development and Mineralization Pattern of The
e Rese tur arc ul h c & a u D q e A v e f l o o Mesa-Rodríguez et al., J Aquac Res Development 2014, 5:6 l p a m n Journal of Aquaculture r e u n o t DOI: 10.4172/2155-9546.1000266 J ISSN: 2155-9546 Research & Development Research Article OpenOpen Access Access Skeletal Development and Mineralization Pattern of the Vertebral Column, Dorsal, Anal and Caudal Fin Complex in Seriola Rivoliana (Valenciennes, 1833) Larvae Mesa-Rodríguez A*, Hernández-Cruz CM, Socorro JA, Fernández-Palacios H, Izquierdo MS and Roo J Aquaculture Research Group, Science and Technology Park, University of Las Palmas de Gran Canaria, P. O. Box 56, 35200 Telde, Canary Islands, Spain Abstract Bone and fins development in Seriola rivoliana were studied from cleared and stained specimens from 3 to 33 days after hatching. The vertebral column began to mineralize in the neural arches at 4.40 ± 0.14 mm Standard Length (SL), continued with the haemal arches and centrums following a cranial-caudal direction. Mineralization of the caudal fin structures started with the caudal rays by 5.12 ± 0.11 mm SL, at the same time that the notochord flexion occurs. The first dorsal and anal fin structures were the hard spines (S), and lepidotrichium (R) by 8.01 ± 0.26 mm SL. The metamorphosis was completed by 11.82 ± 0.4 mm SL. Finally, the fin supports (pterygiophores) and the caudal fins were completely mineralized by 16.1 ± 0.89 mm SL. In addition, the meristic data of 23 structures were provided. -

Appendix 1. (Online Supplementary Material) Species, Gliding Strategies
Appendix 1. (Online Supplementary Material) Species, gliding strategies, species distributions, geographic range sizes, habitat, and egg buoyancy characteristics used for concentrated changes tests. Species Gliding strategy Species distribution (reference #) Geographic range size Habitat (reference #) Egg buoyancy (reference #) Cheilopogon abei (Parin, 1996) 4 wings Indian, Indo-Pacific (1) 2 or more ocean basins meroepipelagic (1) Buoyant (2) Cheilopogon atrisignis (Jenkins, 1903) 4 wings Indian, Pacific (1) 2 or more ocean basins meroepipelgic (3) Buoyant (4) Cheilopogon cyanopterus (Valenciennes, 1847) 4 wings Atlantic, Indo-Pacific (2) 2 or more ocean basins meroepipelgic (3) Non-Buoyant (5) Cheilopogon dorsomacula (Fowler, 1944) 4 wings Pacific (1) within 1 ocean basin holoepipelagic (1) Buoyant (2) Cheilopogon exsiliens (Linnaeus, 1771) 4 wings Atlantic (2) within 1 ocean basin holoepipelagic (3) Buoyant (2,5) Cheilopogon furcatus (Mitchill, 1815) 4 wings Atlantic, Indian, Pacific (6) 2 or more ocean basins holoepipelagic (3) Non-Buoyant (5) Cheilopogon melanurus (Valenciennes, 1847) 4 wings Atlantic (7) within 1 ocean basin meroepipelagic (7) Non-Buoyant (5,8) Cheilopogon pinnatibarbatus (californicus) (Cooper, 1863) 4 wings eastern tropical Pacific (9) within 1 ocean basin meroepipelgic (3) Non-Buoyant (10) Cheilopogon spilonotopterus (Bleeker, 1865) 4 wings Indian and Pacific (1) 2 or more ocean basins meroepipelgic (3) Buoyant (4) Cheilopogon xenopterus (Gilbert, 1890) 4 wings eastern tropical Pacific (11) within 1 ocean basin -

Nocturnal Feeding of Pacific Hake and Jack Mackerel Off the Mouth of the Columbia River, 1998-2004: Implications for Juvenile Salmon Predation Robert L
This article was downloaded by: [Oregon State University] On: 16 August 2011, At: 13:01 Publisher: Taylor & Francis Informa Ltd Registered in England and Wales Registered Number: 1072954 Registered office: Mortimer House, 37-41 Mortimer Street, London W1T 3JH, UK Transactions of the American Fisheries Society Publication details, including instructions for authors and subscription information: http://www.tandfonline.com/loi/utaf20 Nocturnal Feeding of Pacific Hake and Jack Mackerel off the Mouth of the Columbia River, 1998-2004: Implications for Juvenile Salmon Predation Robert L. Emmett a & Gregory K. Krutzikowsky b a Northwest Fisheries Science Center, NOAA Fisheries, 2030 South Marine Science Drive, Newport, Oregon, 97365, USA b Cooperative Institute of Marine Resource Studies, Oregon State University, 2030 South Marine Science Drive, Newport, Oregon, 97365, USA Available online: 09 Jan 2011 To cite this article: Robert L. Emmett & Gregory K. Krutzikowsky (2008): Nocturnal Feeding of Pacific Hake and Jack Mackerel off the Mouth of the Columbia River, 1998-2004: Implications for Juvenile Salmon Predation, Transactions of the American Fisheries Society, 137:3, 657-676 To link to this article: http://dx.doi.org/10.1577/T06-058.1 PLEASE SCROLL DOWN FOR ARTICLE Full terms and conditions of use: http://www.tandfonline.com/page/terms-and- conditions This article may be used for research, teaching and private study purposes. Any substantial or systematic reproduction, re-distribution, re-selling, loan, sub-licensing, systematic supply or distribution in any form to anyone is expressly forbidden. The publisher does not give any warranty express or implied or make any representation that the contents will be complete or accurate or up to date. -

MERLUZA COMÚN Merluccius Gayi Gayi (Guichenot, 1848)
Ficha Pesquera Noviembre - 2008 MERLUZA COMÚN Merluccius gayi gayi (Guichenot, 1848) I. ANTECEDENTES DEL RECURSO Antecedentes biológicos Familia Merlucciidae Orden Gadiformes Clase Actinopterygii Hábitat Batidemersal Alimentación Zooplancton (eufausidos), Necton (peces juveniles), Zoobentos (crustáceos decápodos). Canibalismo Tamaño máximo (cm) 80 cm LT Talla modal 2008 (cm) 35 cm LT (industrial); 35 cm LT (espinel); 37 cm LT (enmalle) Longevidad (años) 15 años Edad de reclutamiento 2-3 años Ciclo de vida El ciclo de vida de esta especie está fuertemente asociado a la columna de agua sobre el área de la plataforma y talud continental de Chile centro-sur (zona nerítica), aunque bajo circunstancias ambientales extraordinarias es posible que ciertos procesos se verifiquen en la zona oceánica aledaña. El ciclo de vida comienza con el desove, el cual se realiza durante todo el año (desovante parcial) aunque el período de mayor intensidad se verifica en invierno-primavera, y un período de desove secundario en febrero- marzo de cada año. Las principales áreas de desove están cercanas a la costa entre Papudo (32º30’ LS) y Bahía San Pedro (40º50’ LS), los huevos desovados son fecundados en el área demersal de la zona nerítica, pasando las larvas a formar parte del necton por un periodo hasta el momento no determinado, y estando sujetas a los típicos procesos de transporte y advección que ocurren a lo largo de la zona centro sur de Chile. Después de un año a un año y medio, los juveniles de merluza común se reclutan al stock, habitando en áreas cercanas a la costa. A partir de los 3 años (34 cm LT), los ejemplares se reclutan a la pesquería y a partir de los 3,5 años (35-37 cm LT) alcanzan la edad de madurez sexual al 50%, constituyéndose en parte del stock adulto, el cual esta asociado a la contracorriente subsuperficial de Chile y Perú (Corriente de Gunther). -

IATTC-94-01 the Tuna Fishery, Stocks, and Ecosystem in the Eastern
INTER-AMERICAN TROPICAL TUNA COMMISSION 94TH MEETING Bilbao, Spain 22-26 July 2019 DOCUMENT IATTC-94-01 REPORT ON THE TUNA FISHERY, STOCKS, AND ECOSYSTEM IN THE EASTERN PACIFIC OCEAN IN 2018 A. The fishery for tunas and billfishes in the eastern Pacific Ocean ....................................................... 3 B. Yellowfin tuna ................................................................................................................................... 50 C. Skipjack tuna ..................................................................................................................................... 58 D. Bigeye tuna ........................................................................................................................................ 64 E. Pacific bluefin tuna ............................................................................................................................ 72 F. Albacore tuna .................................................................................................................................... 76 G. Swordfish ........................................................................................................................................... 82 H. Blue marlin ........................................................................................................................................ 85 I. Striped marlin .................................................................................................................................... 86 J. Sailfish -

Amorphometric and Meristic Study of the Halfbeak, Hyporhamphus
W&M ScholarWorks Dissertations, Theses, and Masters Projects Theses, Dissertations, & Master Projects 1993 Amorphometric and Meristic Study of the Halfbeak, Hyporhamphus unifasciatus (Teleostei: Hemiramphidae) from the Western Atlantic, with the Description of a New Species Heidi M. Banford College of William and Mary - Virginia Institute of Marine Science Follow this and additional works at: https://scholarworks.wm.edu/etd Part of the Marine Biology Commons, and the Oceanography Commons Recommended Citation Banford, Heidi M., "Amorphometric and Meristic Study of the Halfbeak, Hyporhamphus unifasciatus (Teleostei: Hemiramphidae) from the Western Atlantic, with the Description of a New Species" (1993). Dissertations, Theses, and Masters Projects. Paper 1539617658. https://dx.doi.org/doi:10.25773/v5-pbsc-sy52 This Thesis is brought to you for free and open access by the Theses, Dissertations, & Master Projects at W&M ScholarWorks. It has been accepted for inclusion in Dissertations, Theses, and Masters Projects by an authorized administrator of W&M ScholarWorks. For more information, please contact [email protected]. A MORPHOMETRIC AND MERISTIC STUDY OF THE HALFBEAK, HYPORHAMPHUS UNIFASCIATUS (TELEOSTEI: HEMIRAMPHIDAE) FROM THE WESTERN ATLANTIC, WITH THE DESCRIPTION OF A NEW SPECIES A Thesis Presented to The Faculty of the School of Marine Science The College of William and Mary in Virginia In Partial Fulfillment Of the Requirements for the Degree of Master of Arts by Heidi M. Banford 1993 This thesis is submitted in partial fulfillment of the requirements for the degree of Master of Arts Heidi M. Banford Approved, July 1993 Jojm A. Musick,' Ph.D. flmittee Chairman/Advisor ~ t M . ^ Herbert M. Austin, Ph.D. -

HAKES of the WORLD (Family Merlucciidae)
ISSN 1020-8682 FAO Species Catalogue for Fishery Purposes No. 2 HAKES OF THE WORLD (Family Merlucciidae) AN ANNOTATED AND ILLUSTRATED CATALOGUE OF HAKE SPECIES KNOWN TO DATE FAO Species Catalogue for Fishery Purposes No. 2 FIR/Cat. 2 HAKES OF THE WORLD (Family Merlucciidae) AN ANNOTATED AND ILLUSTRATED CATALOGUE OF HAKE SPECIES KNOWN TO DATE by D. Lloris Instituto de Ciencias del Mar (CMIMA-CSIC) Barcelona, Spain J. Matallanas Facultad de Ciencias Universidad Autónoma de Barcelona Bellaterra, Barcelona, Spain and P. Oliver Instituto Español de Oceanografia Palma de Mallorca, Spain FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS Rome, 2005 The designations employed and the presentation of material in this information product do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organization of the United Nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitation of its frontiers or boundaries. ISBN 92-5-104984-X All rights reserved. Reproduction and dissemination of material in this information product for educational or other non-commercial purposes are authorized without any prior written permission from the copyright holders provided the source is fully acknowledged. Reproduction of material in this information product for resale or other commercial purposes is prohibited without written permission of the copyright holders. Applications for such permission should be addressed to the Chief, Publishing Management Service, Information Division, FAO, Viale delle Terme di Caracalla, 00100 Rome, Italy by e-mail to [email protected] © FAO 2005 Hakes of the World iii PREPARATION OF THIS DOCUMENT his catalogue was prepared under the FAO Fisheries Department Regular Programme by the Species Identification and TData Programme in the Marine Resources Service of the Fishery Resources Division. -
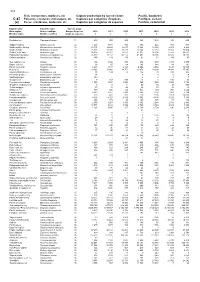
Fish, Crustaceans, Molluscs, Etc Capture Production by Species
534 Fish, crustaceans, molluscs, etc Capture production by species items Pacific, Southeast C-87 Poissons, crustacés, mollusques, etc Captures par catégories d'espèces Pacifique, sud-est (a) Peces, crustáceos, moluscos, etc Capturas por categorías de especies Pacífico, sudoriental English name Scientific name Species group Nom anglais Nom scientifique Groupe d'espèces 2010 2011 2012 2013 2014 2015 2016 Nombre inglés Nombre científico Grupo de especies t t t t t t t Flatfishes nei Pleuronectiformes 31 613 302 806 323 1 804 483 693 Tadpole codling Salilota australis 32 1 400 1 091 768 374 522 703 536 Southern blue whiting Micromesistius australis 32 23 301 19 629 16 675 15 304 10 036 8 809 8 269 Southern hake Merluccius australis 32 25 361 20 909 20 288 19 346 12 393 16 150 16 804 South Pacific hake Merluccius gayi 32 90 305 82 977 72 872 92 031 96 196 77 283 98 662 Patagonian grenadier Macruronus magellanicus 32 74 330 70 137 62 175 47 602 39 138 37 475 28 108 Chilean grenadier Coelorinchus chilensis 32 156 134 136 91 54 59 47 Sea catfishes nei Ariidae 33 406 1 426 852 876 1 217 1 185 1 285 Snake eels nei Ophichthidae 33 38 65 114 142 144 49 181 Pacific cornetfish Fistularia corneta 33 6 443 4 513 4 323 4 854 2 534 7 630 19 559 Mullets nei Mugilidae 33 10 821 13 400 18 751 13 876 14 290 14 044 17 345 Snooks(=Robalos) nei Centropomus spp 33 98 104 78 136 79 310 305 Broomtail grouper Mycteroperca xenarcha 33 14 .. -

© Iccat, 2007
A5 By-catch Species APPENDIX 5: BY-CATCH SPECIES A.5 By-catch species By-catch is the unintentional/incidental capture of non-target species during fishing operations. Different types of fisheries have different types and levels of by-catch, depending on the gear used, the time, area and depth fished, etc. Article IV of the Convention states: "the Commission shall be responsible for the study of the population of tuna and tuna-like fishes (the Scombriformes with the exception of Trichiuridae and Gempylidae and the genus Scomber) and such other species of fishes exploited in tuna fishing in the Convention area as are not under investigation by another international fishery organization". The following is a list of by-catch species recorded as being ever caught by any major tuna fishery in the Atlantic/Mediterranean. Note that the lists are qualitative and are not indicative of quantity or mortality. Thus, the presence of a species in the lists does not imply that it is caught in significant quantities, or that individuals that are caught necessarily die. Skates and rays Scientific names Common name Code LL GILL PS BB HARP TRAP OTHER Dasyatis centroura Roughtail stingray RDC X Dasyatis violacea Pelagic stingray PLS X X X X Manta birostris Manta ray RMB X X X Mobula hypostoma RMH X Mobula lucasana X Mobula mobular Devil ray RMM X X X X X Myliobatis aquila Common eagle ray MYL X X Pteuromylaeus bovinus Bull ray MPO X X Raja fullonica Shagreen ray RJF X Raja straeleni Spotted skate RFL X Rhinoptera spp Cownose ray X Torpedo nobiliana Torpedo -

Among the World's Most Popular Game Fishes, Tunas Are Also
ÜBER-FISH Among the World’s Most Popular Game Fishes, Tunas Are Also Some of the Most Highly Evolved and Sophisticated of All the Ocean’s Predators BY DOUG OLANDER DANIEL GOEZ DANIEL 74 DECEMBER 2017 SPORTFISHINGMAG.COM 75 The Family Tree minimizes drag with a very low reduce the turbulence in the Tunas are part of the family drag coefficient,” optimizing effi- water ahead of the tail. Scombridae, which also includes cient swimming both at cruise Unlike most fishes with broad, mackerels, large and small. But and burst. While most fishes bend flexible tails that bend to scoop there are tunas, and then there their bodies side to side when water to move a fish forward, are, well, “true tunas.” moving forward, tunas’ bodies tunas derive tremendous That is, two groups don’t bend. They’re essentially thrust with thin, hard, lunate WHILE MOST FISHES BEND ( sometimes known as “tribes”) rigid, solid torpedoes. ( crescent-moon-shaped) tails dominate the tuna clan. One is And these torpedoes are that beat constantly, capable of THEIR BODIES SIDE TO SIDE Thunnini, which is the group perfectly streamlined, their 10 to 12 or more beats per second. considered true tunas, charac- larger fins fitting perfectly into That relentless thrust accounts WHEN MOVING FORWARD, terized by two separate dorsal grooves so no part of these fins for the unstoppable runs that fins and a relatively thick body. a number of highly specialized protrudes above the body surface. tuna make repeatedly when TUNAS’ BODIES DON’T BEND. The 15 species of Thunnini are features facilitate these They lack the convex eyes of hooked. -

Descriptions of Euthynnus and Auxis Larvae from the Pacific and Atlantic Oceans and Adjacent Seas
library THE GARLSBERG FOUNDATION’S OCEANOGRAPHIGAL EXPEDITION ROUND THE WORLD 1928—30 AND PREVIOUS “DANA”-EXPEDITIONS UNDER THE LEADERSHIP OF THE LATE PROFESSOR JOHANNES SCHMIDT DANA-BEPOBT No. 50. DESCRIPTIONS OF EUTHYNNUS AND AUXIS LARVAE FROM THE PACIFIC AND ATLANTIC OCEANS AND ADJACENT SEAS BY WALTER M. MATSUMOTO U.S. FISH AND WILDLIFE SERVICE WITH 31 FIGURES IN THE TEXT PUBLISHED BY THE CARLSBERG FOUNDATION THIS PAPER MAY BE EEFEEBED TO AS: •DANA-REPOKT No. 50, 1959« COPENHAGEN ANDR. FRED. H0ST A S 0 N PRINTED BY BIANCO LUNO A/S CONTENTS Page Introduction ...................................................................... 3 Descriptions of larvae and postlarvae................. 21 Methods.............................................................................. 4 Auxis type I ......................................................... 21 Genus E uthynnus.............................................................. 5 Auxis type I I ....................................................... 2:i Notes on adults and juveniles ............................. 5 Discussion of species dilTerences........................... 25 Descriptions of larvae and postlarvae................. 7 Geographical distribution of Euthynnus and Auxis Euthynnus tineatus.............................................. 7 larvae............................................................................ 25 Euthynnus alletteratus.......................................... 11 Spawning areas as indicated by larval catches___ 27 Euthynnus ijaito .................................................. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences.