Human T-Cell Lymphotropic Virus Type 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Food Handler Exclusion Guidelines
Food Handler Exclusion Guidelines A guide for determining suitable exclusion periods for ill food handlers November 2017 Food handler exclusion guidelines Published by the State of Queensland (Queensland Health), November 2017 This document is licensed under a Creative Commons Attribution 3.0 Australia licence. To view a copy of this licence, visit creativecommons.org/licenses/by/3.0/au © State of Queensland (Queensland Health) 2017 You are free to copy, communicate and adapt the work, as long as you attribute the State of Queensland (Queensland Health). For more information contact: Food Safety Standards and Regulation, Department of Health, GPO Box 48, Brisbane QLD 4001, email [email protected], phone 07 3328 9310. An electronic version of this document is available at www.health.qld.gov.au. Disclaimer: The content presented in this publication is distributed by the Queensland Government as an information source only. The State of Queensland makes no statements, representations or warranties about the accuracy, completeness or reliability of any information contained in this publication. The State of Queensland disclaims all responsibility and all liability (including without limitation for liability in negligence) for all expenses, losses, damages and costs you might incur as a result of the information being inaccurate or incomplete in any way, and for any reason reliance was placed on such information. Food Handler Exclusion Guidelines - ii - Contents 1. Introduction ............................................................................................... -

PEDIARIX Is a Vaccine
HIGHLIGHTS OF PRESCRIBING INFORMATION • If Guillain-Barré syndrome occurs within 6 weeks of receipt of a prior These highlights do not include all the information needed to use vaccine containing tetanus toxoid, the decision to give PEDIARIX should PEDIARIX safely and effectively. See full prescribing information for be based on potential benefits and risks. (5.2) PEDIARIX. • The tip caps of the prefilled syringes contain natural rubber latex which may cause allergic reactions. (5.3) PEDIARIX [Diphtheria and Tetanus Toxoids and Acellular Pertussis • Syncope (fainting) can occur in association with administration of Adsorbed, Hepatitis B (Recombinant) and Inactivated Poliovirus injectable vaccines, including PEDIARIX. Procedures should be in place Vaccine], Suspension for Intramuscular Injection to avoid falling injury and to restore cerebral perfusion following Initial U.S. Approval: 2002 syncope. (5.4) • If temperature ≥105°F, collapse or shock-like state, or persistent, ----------------------------- INDICATIONS AND USAGE ---------------------------- PEDIARIX is a vaccine indicated for active immunization against diphtheria, inconsolable crying lasting ≥3 hours have occurred within 48 hours after tetanus, pertussis, infection caused by all known subtypes of hepatitis B virus, receipt of a pertussis-containing vaccine, or if seizures have occurred and poliomyelitis. PEDIARIX is approved for use as a 3-dose series in infants within 3 days after receipt of a pertussis-containing vaccine, the decision born of hepatitis B surface antigen (HBsAg)-negative mothers. PEDIARIX to give PEDIARIX should be based on potential benefits and risks. (5.5) may be given as early as 6 weeks of age through 6 years of age (prior to the • For children at higher risk for seizures, an antipyretic may be 7th birthday). -
Diphtheria, Tetanus, Pertussis, Polio, Hib and Hepatitis B Vaccine for Babies and Children
Diphtheria, Tetanus, Pertussis, Polio, Hib and Hepatitis B vaccine for babies and children This leaflet tells you about the DTaP/IPV/Hib/ HepB vaccine, also known as “6 in 1” as it protects against six diseases, diphtheria, tetanus, pertussis (whooping cough), polio, Haemophilus influenzae type b and hepatitis B (HepB) disease. What does the vaccine protect against? Diphtheria Diphtheria is a serious disease that usually begins with a sore throat and can quickly cause breathing problems. It can damage the heart and nervous system and, in severe cases, can kill. Before diphtheria vaccine was introduced in the UK, there were up to 70,000 cases of diphtheria and around 5,000 deaths a year. Diphtheria can be spread from person to person through close contact. Tetanus Tetanus is a disease affecting the nervous system which can cause muscle spasms and breathing problems and can kill. It is caused when germs found in soil and manure get into the body through wounds or burns. Tetanus cannot be passed from person to person. 2 Published June 2018 Pertussis (whooping cough) Whooping cough is a disease that can cause long bouts of coughing and choking, making it hard to breathe. It can last for up to 10 weeks and babies under one year of age are most at risk. The disease is very serious and can kill. Before the pertussis vaccine was introduced, the average number of cases reported each year in the UK was 120,000 and 92 children died in the year before the vaccine was introduced. Whooping cough is usually spread by coughs and sneezes. -

Hepatitis B? HEPATITIS B Hepatitis B Is a Contagious Liver Disease That Results from Infection with the Hepatitis B Virus
What is Hepatitis B? HEPATITIS B Hepatitis B is a contagious liver disease that results from infection with the Hepatitis B virus. When first infected, a person can develop Are you at risk? an “acute” infection, which can range in severity from a very mild illness with few or no symptoms to a serious condition requiring hospitalization. Acute Hepatitis B refers to the first 6 months after someone is exposed to the Hepatitis B virus. Some people are able to fight the infection and clear the virus. For others, the infection remains and leads to a “chronic,” or lifelong, illness. Chronic Hepatitis B refers to the illness that occurs when the Hepatitis B virus remains in a person’s body. Over time, the infection can cause serious health problems. How is Hepatitis B spread? Hepatitis B is usually spread when blood, semen, or other body fluids from a person infected with the Hepatitis B virus enter the body of someone who is not infected. This can happen through having sex with an infected partner; sharing needles, syringes, or other injection drug equipment; or from direct contact with the blood or open sores of an infected person. Hepatitis B can also be passed from an infected mother to her baby at birth. Who should be tested for Hepatitis B? Approximately 1.2 million people in the United States and 350 million people worldwide have Hepatitis B. Testing for Hepatitis B is recommended for certain groups of people, including: Most are unaware of their infection. ■ People born in Asia, Africa, and other regions with moderate or high rates Is Hepatitis B common? of Hepatitis B (see map) Yes. -
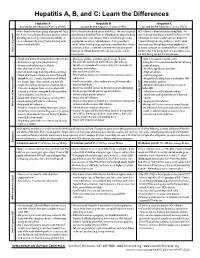
Hepatitis A, B, and C: Learn the Differences
Hepatitis A, B, and C: Learn the Differences Hepatitis A Hepatitis B Hepatitis C caused by the hepatitis A virus (HAV) caused by the hepatitis B virus (HBV) caused by the hepatitis C virus (HCV) HAV is found in the feces (poop) of people with hepa- HBV is found in blood and certain body fluids. The virus is spread HCV is found in blood and certain body fluids. The titis A and is usually spread by close personal contact when blood or body fluid from an infected person enters the body virus is spread when blood or body fluid from an HCV- (including sex or living in the same household). It of a person who is not immune. HBV is spread through having infected person enters another person’s body. HCV can also be spread by eating food or drinking water unprotected sex with an infected person, sharing needles or is spread through sharing needles or “works” when contaminated with HAV. “works” when shooting drugs, exposure to needlesticks or sharps shooting drugs, through exposure to needlesticks on the job, or from an infected mother to her baby during birth. or sharps on the job, or sometimes from an infected How is it spread? Exposure to infected blood in ANY situation can be a risk for mother to her baby during birth. It is possible to trans- transmission. mit HCV during sex, but it is not common. • People who wish to be protected from HAV infection • All infants, children, and teens ages 0 through 18 years There is no vaccine to prevent HCV. -

Hepatitis B Vaccine – Frequently Asked Questions (Information from the CDC)
AAMC Standardized Immunization Form 2020 Hepatitis B Vaccine – Frequently Asked Questions (Information from the CDC) 1. What are the hepatitis B vaccines licensed for use in the United States? Three single-antigen vaccines and two combination vaccines are currently licensed in the United States. Single-antigen hepatitis B vaccines: • ENGERIX-B® • RECOMBIVAX HB® • HEPLISAV-B™ Combination vaccines: • PEDIARIX®: Combined hepatitis B, diphtheria, tetanus, acellular pertussis (DTaP), and inactivated poliovirus (IPV) vaccine. Cannot be administered before age 6 weeks or after age 7 years. • TWINRIX®: Combined Hepatitis A and hepatitis B vaccine. Recommended for people aged ≥18 years who are at increased risk for both HAV and HBV infections. 2. What are the recommended schedules for hepatitis B vaccination? The vaccination schedule most often used for children and adults is three doses given at 0, 1, and 6 months. Alternate schedules have been approved for certain vaccines and/or populations. A new formulation, Heplisav-B (HepB-CpG), is approved to be given as two doses one month apart. 3. If there is an interruption between doses of hepatitis B vaccine, does the vaccine series need to be restarted? No. The series does not need to be restarted but the following should be considered: • If the vaccine series was interrupted after the first dose, the second dose should be administered as soon as possible. • The second and third doses should be separated by an interval of at least 8 weeks. • If only the third dose is delayed, it should be administered as soon as possible. 4. Is it harmful to administer an extra dose of hepatitis B vaccine or to repeat the entire vaccine series if documentation of the vaccination history is unavailable or the serology test is negative? No, administering extra doses of single-antigen hepatitis B vaccine is not harmful. -

Package Inserts
Individuals using assistive technology may not be able to fully access the information contained in this file. For assistance, please send an e-mail to: [email protected] and include 508 Accommodation and the title of the document in the subject line of your e-mail. HIGHLIGHTS OF PRESCRIBING INFORMATION -----------------------WARNINGS AND PRECAUTIONS------------------------ These highlights do not include all the information needed to use • Carefully consider benefits and risks before administering VAXELIS to VAXELIS safely and effectively. See full prescribing information for persons with a history of: VAXELIS. - fever ≥40.5°C (≥105°F), hypotonic-hyporesponsive episode (HHE) or VAXELISTM (Diphtheria and Tetanus Toxoids and Acellular Pertussis, persistent, inconsolable crying lasting ≥3 hours within 48 hours after a Inactivated Poliovirus, Haemophilus b Conjugate and Hepatitis B previous pertussis-containing vaccine. (5.2) Vaccine) - seizures within 3 days after a previous pertussis-containing vaccine. (5.2) Suspension for Intramuscular Injection Initial U.S. Approval: 2018 • If Guillain-Barré syndrome occurred within 6 weeks of receipt of a prior vaccine containing tetanus toxoid, the risk for Guillain-Barré syndrome ----------------------------RECENT MAJOR CHANGES ------------------------ may be increased following VAXELIS. (5.3) Dosage and Administration (2.2) 09/2020 • Apnea following intramuscular vaccination has been observed in some ----------------------------INDICATIONS AND USAGE--------------------------- infants born prematurely. The decision about when to administer an VAXELIS is a vaccine indicated for active immunization to prevent intramuscular vaccine, including VAXELIS, to an infant born prematurely diphtheria, tetanus, pertussis, poliomyelitis, hepatitis B, and invasive disease should be based on consideration of the individual infant’s medical status due to Haemophilus influenzae type b. -

Recommended Adult Immunization Schedule
Recommended Adult Immunization Schedule UNITED STATES for ages 19 years or older 2021 Recommended by the Advisory Committee on Immunization Practices How to use the adult immunization schedule (www.cdc.gov/vaccines/acip) and approved by the Centers for Disease Determine recommended Assess need for additional Review vaccine types, Control and Prevention (www.cdc.gov), American College of Physicians 1 vaccinations by age 2 recommended vaccinations 3 frequencies, and intervals (www.acponline.org), American Academy of Family Physicians (www.aafp. (Table 1) by medical condition and and considerations for org), American College of Obstetricians and Gynecologists (www.acog.org), other indications (Table 2) special situations (Notes) American College of Nurse-Midwives (www.midwife.org), and American Academy of Physician Assistants (www.aapa.org). Vaccines in the Adult Immunization Schedule* Report y Vaccines Abbreviations Trade names Suspected cases of reportable vaccine-preventable diseases or outbreaks to the local or state health department Haemophilus influenzae type b vaccine Hib ActHIB® y Clinically significant postvaccination reactions to the Vaccine Adverse Event Hiberix® Reporting System at www.vaers.hhs.gov or 800-822-7967 PedvaxHIB® Hepatitis A vaccine HepA Havrix® Injury claims Vaqta® All vaccines included in the adult immunization schedule except pneumococcal 23-valent polysaccharide (PPSV23) and zoster (RZV) vaccines are covered by the Hepatitis A and hepatitis B vaccine HepA-HepB Twinrix® Vaccine Injury Compensation Program. Information on how to file a vaccine injury Hepatitis B vaccine HepB Engerix-B® claim is available at www.hrsa.gov/vaccinecompensation. Recombivax HB® Heplisav-B® Questions or comments Contact www.cdc.gov/cdc-info or 800-CDC-INFO (800-232-4636), in English or Human papillomavirus vaccine HPV Gardasil 9® Spanish, 8 a.m.–8 p.m. -

Vaccinations for Adults with Chronic Liver Disease Or Infection
Vaccinations for Adults with Chronic Liver Disease or Infection This table shows which vaccinations you should have to protect your health if you have chronic hepatitis B or C infection or chronic liver disease (e.g., cirrhosis). Make sure you and your healthcare provider keep your vaccinations up to date. Vaccine Do you need it? Hepatitis A Yes! Your chronic liver disease or infection puts you at risk for serious complications if you get infected with the (HepA) hepatitis A virus. If you’ve never been vaccinated against hepatitis A, you need 2 doses of this vaccine, usually spaced 6–18 months apart. Hepatitis B Yes! If you already have chronic hepatitis B infection, you won’t need hepatitis B vaccine. However, if you have (HepB) hepatitis C or other causes of chronic liver disease, you do need hepatitis B vaccine. The vaccine is given in 2 or 3 doses, depending on the brand. Ask your healthcare provider if you need screening blood tests for hepatitis B. Hib (Haemophilus Maybe. Some adults with certain high-risk conditions, for example, lack of a functioning spleen, need vaccination influenzae type b) with Hib. Talk to your healthcare provider to find out if you need this vaccine. Human Yes! You should get this vaccine if you are age 26 years or younger. Adults age 27 through 45 may also be vacci- papillomavirus nated against HPV after a discussion with their healthcare provider. The vaccine is usually given in 3 doses over a (HPV) 6-month period. Influenza Yes! You need a dose every fall (or winter) for your protection and for the protection of others around you. -

Transmission of Hepatitis B, Hepatitis C and Human Immunodeficiency Viruses Through Unsafe Injections in the Developing World: Model-Based Regional Estimates A
Transmission of hepatitis B, hepatitis C and human immunodeficiency viruses through unsafe injections in the developing world: model-based regional estimates A. Kane,1 J. Lloyd,2 M. Zaffran,3 L. Simonsen,4 & M. Kane5 Thousands of millions of injections are delivered every year in developing countries, many of them unsafe, and the transmission of certain bloodborne pathogens via this route is thought to be a major public health problem. In this article we report global and regional estimates of the number of hepatitis B virus (HBV), hepatitis Cvirus (HCV)and human immunodeficiency virus (HIV) infections that may occur from unsafe injections in the developing world. The estimates were determined using quantitative data on unsafe injection practices, transmission efficiency and disease burden of HBV, HCV and HIV and the prevalence of injection use obtained from a review of the literature. A simple mass-action model was used consisting of a generalized linear equation with variables accounting for the prevalence of a pathogen in a population, susceptibility of a population, transmission efficiency of the pathogen, proportion of injections that are unsafe, and the number of injections received. The model was applied to world census data to generate conservative estimates of incidence of transmission of bloodborne pathogens that may be attributable to unsafe injections. The model suggests that approximately 8±16 million HBV, 2.3±4.7 million HCV and 80 000±160 000 HIV infections may result every year from unsafe injections. The estimated range for HBV infections is in accordance with several epidemiological studies that attributed at least 20% of all new HBV infections to unsafe injections in developing countries. -

Hepatitis B Factsheet: COVID-19 and Hep B
Hepatitis B factsheet: COVID-19 and hep B For more information about anything in this factsheet, phone the Hepatitis Infoline on 1800 803 990 or go to www.hep.org.au COVID-19 and hep B Frequently Asked Questions (FAQ) There’s so much information swirling around these days as we deal with the changing world around us due to COVID-19. There’s lots of incorrect information and it’s easy to be overwhelmed and confused. On this page, we’ll try to clear some things up. It is important that information is helpful, clear, and checked and confirmed by experts. Please share the following information – it comes from health and medical specialists. COVID-19 is an acronym that stands for COronaVIrus Disease 2019. You might hear it called coronavirus, corona, SARS-CoV-2, or even rona. We’ll call it COVID-19 or coronavirus in this FAQ but all these terms are essentially talking about the same thing and we’ll explain them at the bottom. You’ll also hear new or unfamiliar terms like “social distancing,” “physical distancing,” “self-isolation,” and “quarantine”. These terms and what they actually mean can become confusing so we’ve including a glossary at the bottom of this FAQ. This virus has only been known about since around December 2019 so it’s all very new, information changes quickly, and we’re all learning day-by-day and week-by-week. Compare COVID-19 to hep B which we’ve known about since the 1980s and hep B even earlier in the 1960s! We’ll regularly update this page as new information comes to light. -
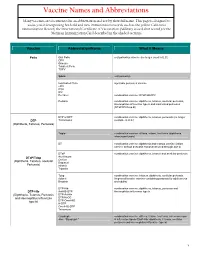
Vaccine Names and Abbreviations Vaccine Names and Abbreviations Many Vaccines Are Documented in an Abbreviation and Not by Their Full Name
Vaccine Names and Abbreviations Vaccine Names and Abbreviations Many vaccines are documented in an abbreviation and not by their full name. This page is designed to assist you in interpreting both old and new immunization records such as the yellow California Immunization Record, the International Certificate of Vaccination (Military issued shot record) or the Mexican Immunization Card described in the shaded sections. Vaccine Abbreviation/Name What it Means Polio Oral Polio oral poliovirus vaccine (no longer used in U.S.) OPV Orimune Trivalent Polio TOPV Sabin oral poliovirus Inactivated Polio injectable poliovirus vaccine eIPV IPOL IPV Pentacel combination vaccine: DTaP/Hib/IPV Pediarix combination vaccine: diphtheria, tetanus, acellular pertussis, Haemophilus influenzae type b and inactivated poliovirus (DTaP/IPV/Hep B) DTP or DPT combination vaccine: diphtheria, tetanus, pertussis (no longer DTP Tri-Immunol available in U.S.) (Diphtheria, Tetanus, Pertussis) Triple combination vaccine: difteria, tétano, tos ferina (diphtheria, tetanus pertussis) DT combination vaccine: diphtheria and tetanus vaccine (infant vaccine without pertussis component used through age 6) DTaP combination vaccine: diphtheria, tetanus and acellular pertussis DTaP/Tdap Acel-Imune Certiva (Diphtheria, Tetanus, acellular Daptacel Pertussis) Infanrix Tripedia Tdap combination vaccine: tetanus, diphtheria, acellular pertussis Adacel (improved booster vaccine containing pertussis for adolescents Boostrix and adults) DTP-Hib combination vaccine: diphtheria, tetanus,