Neuromuscular Junction: a Dissertation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Oligodendrocytes in Development, Myelin Generation and Beyond
cells Review Oligodendrocytes in Development, Myelin Generation and Beyond Sarah Kuhn y, Laura Gritti y, Daniel Crooks and Yvonne Dombrowski * Wellcome-Wolfson Institute for Experimental Medicine, Queen’s University Belfast, Belfast BT9 7BL, UK; [email protected] (S.K.); [email protected] (L.G.); [email protected] (D.C.) * Correspondence: [email protected]; Tel.: +0044-28-9097-6127 These authors contributed equally. y Received: 15 October 2019; Accepted: 7 November 2019; Published: 12 November 2019 Abstract: Oligodendrocytes are the myelinating cells of the central nervous system (CNS) that are generated from oligodendrocyte progenitor cells (OPC). OPC are distributed throughout the CNS and represent a pool of migratory and proliferative adult progenitor cells that can differentiate into oligodendrocytes. The central function of oligodendrocytes is to generate myelin, which is an extended membrane from the cell that wraps tightly around axons. Due to this energy consuming process and the associated high metabolic turnover oligodendrocytes are vulnerable to cytotoxic and excitotoxic factors. Oligodendrocyte pathology is therefore evident in a range of disorders including multiple sclerosis, schizophrenia and Alzheimer’s disease. Deceased oligodendrocytes can be replenished from the adult OPC pool and lost myelin can be regenerated during remyelination, which can prevent axonal degeneration and can restore function. Cell population studies have recently identified novel immunomodulatory functions of oligodendrocytes, the implications of which, e.g., for diseases with primary oligodendrocyte pathology, are not yet clear. Here, we review the journey of oligodendrocytes from the embryonic stage to their role in homeostasis and their fate in disease. We will also discuss the most common models used to study oligodendrocytes and describe newly discovered functions of oligodendrocytes. -

Neuregulin 1–Erbb2 Signaling Is Required for the Establishment of Radial Glia and Their Transformation Into Astrocytes in Cerebral Cortex
Neuregulin 1–erbB2 signaling is required for the establishment of radial glia and their transformation into astrocytes in cerebral cortex Ralf S. Schmid*, Barbara McGrath*, Bridget E. Berechid†, Becky Boyles*, Mark Marchionni‡, Nenad Sˇ estan†, and Eva S. Anton*§ *University of North Carolina Neuroscience Center and Department of Cell and Molecular Physiology, University of North Carolina School of Medicine, Chapel Hill, NC 27599; †Department of Neurobiology, Yale University School of Medicine, New Haven, CT 06510; and ‡CeNes Pharamceuticals, Inc., Norwood, MA 02062 Communicated by Pasko Rakic, Yale University School of Medicine, New Haven, CT, January 27, 2003 (received for review December 12, 2002) Radial glial cells and astrocytes function to support the construction mine whether NRG-1-mediated signaling is involved in radial and maintenance, respectively, of the cerebral cortex. However, the glial cell development and differentiation in the cerebral cortex. mechanisms that determine how radial glial cells are established, We show that NRG-1 signaling, involving erbB2, may act in maintained, and transformed into astrocytes in the cerebral cortex are concert with Notch signaling to exert a critical influence in the not well understood. Here, we show that neuregulin-1 (NRG-1) exerts establishment, maintenance, and appropriate transformation of a critical role in the establishment of radial glial cells. Radial glial cell radial glial cells in cerebral cortex. generation is significantly impaired in NRG mutants, and this defect can be rescued by exogenous NRG-1. Down-regulation of expression Materials and Methods and activity of erbB2, a member of the NRG-1 receptor complex, leads Clonal Analysis to Study NRG’s Role in the Initial Establishment of to the transformation of radial glial cells into astrocytes. -

Pre-Oligodendrocytes from Adult Human CNS
The Journal of Neuroscience, April 1992, 12(4): 1538-l 547 Pre-Oligodendrocytes from Adult Human CNS Regina C. Armstrong,lJ Henry H. Dorn, l,b Conrad V. Kufta,* Emily Friedman,3 and Monique E. Dubois-Dalcq’ ‘Laboratory of Viral and Molecular Pathogenesis, and %urgical Neurology Branch, National Institute of Neurological Disorders and Stroke, Bethesda, Maryland 20892 and 3Department of Neurosurgery, University of Pennsylvania, Philadelphia, Pennsylvania 19104-3246 CNS remyelination and functional recovery often occur after Rapid and efficient neurotransmission is dependent upon the experimental demyelination in adult rodents. This has been electrical insulating capacity of the myelin sheath around axons attributed to the ability of mature oligodendrocytes and/or (reviewed in Ritchie, 1984a,b). Nerve conduction is impaired their precursor cells to divide and regenerate in response after loss of the myelin sheath and results in severe neurological to signals in demyelinating lesions. To determine whether dysfunction in human demyelinating diseases such as multiple oligodendrocyte precursor cells exist in the adult human sclerosis (MS). Remyelination can occur in the CNS of MS CNS, we have cultured white matter from patients under- patients but appears to be limited (Perier and Gregoire, 1965; going partial temporal lobe resection for intractable epilep- Prineas et al., 1984). Studies of acute MS cases have revealed sy. These cultures contained a population of process-bear- that recent demyelinating lesions can exhibit remyelination that ing cells that expressed antigens recognized by the 04 appears to correlate with the generation of new oligodendrocytes monoclonal antibody, but these cells did not express galac- (Prineas et al., 1984; Raine et al., 1988). -
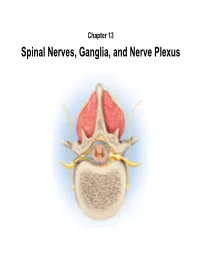
Spinal Nerves, Ganglia, and Nerve Plexus Spinal Nerves
Chapter 13 Spinal Nerves, Ganglia, and Nerve Plexus Spinal Nerves Posterior Spinous process of vertebra Posterior root Deep muscles of back Posterior ramus Spinal cord Transverse process of vertebra Posterior root ganglion Spinal nerve Anterior ramus Meningeal branch Communicating rami Anterior root Vertebral body Sympathetic ganglion Anterior General Anatomy of Nerves and Ganglia • Spinal cord communicates with the rest of the body by way of spinal nerves • nerve = a cordlike organ composed of numerous nerve fibers (axons) bound together by connective tissue – mixed nerves contain both afferent (sensory) and efferent (motor) fibers – composed of thousands of fibers carrying currents in opposite directions Anatomy of a Nerve Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Epineurium Perineurium Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Endoneurium Nerve Rootlets fiber Posterior root Fascicle Posterior root ganglion Anterior Blood root vessels Spinal nerve (b) Copyright by R.G. Kessel and R.H. Kardon, Tissues and Organs: A Text-Atlas of Scanning Electron Microscopy, 1979, W.H. Freeman, All rights reserved Blood vessels Fascicle Epineurium Perineurium Unmyelinated nerve fibers Myelinated nerve fibers (a) Endoneurium Myelin General Anatomy of Nerves and Ganglia • nerves of peripheral nervous system are ensheathed in Schwann cells – forms neurilemma and often a myelin sheath around the axon – external to neurilemma, each fiber is surrounded by -

The Peripheral Nervous System
The Peripheral Nervous System Dr. Ali Ebneshahidi Peripheral Nervous System (PNS) – Consists of 12 pairs of cranial nerves and 31 pairs of spinal nerves. – Serves as a critical link between the body and the central nervous system. – peripheral nerves contain an outermost layer of fibrous connective tissue called epineurium which surrounds a thinner layer of fibrous connective tissue called perineurium (surrounds the bundles of nerve or fascicles). Individual nerve fibers within the nerve are surrounded by loose connective tissue called endoneurium. Cranial Nerves Cranial nerves are direct extensions of the brain. Only Nerve I (olfactory) originates from the cerebrum, the remaining 11 pairs originate from the brain stem. Nerve I (Olfactory)- for the sense of smell (sensory). Nerve II (Optic)- for the sense of vision (sensory). Nerve III (Oculomotor)- for controlling muscles and accessory structures of the eyes ( primarily motor). Nerve IV (Trochlear)- for controlling muscles of the eyes (primarily motor). Nerve V (Trigeminal)- for controlling muscles of the eyes, upper and lower jaws and tear glands (mixed). Nerve VI (Abducens)- for controlling muscles that move the eye (primarily motor). Nerve VII (Facial) – for the sense of taste and controlling facial muscles, tear glands and salivary glands (mixed). Nerve VIII (Vestibulocochlear)- for the senses of hearing and equilibrium (sensory). Nerve IX (Glossopharyngeal)- for controlling muscles in the pharynx and to control salivary glands (mixed). Nerve X (Vagus)- for controlling muscles used in speech, swallowing, and the digestive tract, and controls cardiac and smooth muscles (mixed). Nerve XI (Accessory)- for controlling muscles of soft palate, pharynx and larynx (primarily motor). Nerve XII (Hypoglossal) for controlling muscles that move the tongue ( primarily motor). -
PN1 (Midha) Microanatomy of Peripheral Nerves-Part1.Pdf
Peripheral Nerve Basics • Consist of processes of cell bodies found in the DRG, anterior horn, and autonomic ganglia • Organized by several distinct connective tissue layers – the epineurium, perineurium, and endoneurium • Vascular supply provided by the vasa nervorum Peripheral Nerve Basics • Neuronal processes bound into fascicles by perineurium • Fascicles bound into nerves by epineurium • Endoneurium is a division of the perineurium which form thin layers of connective tissue surrounding neuronal fibers in a fascicle Sural nerve in cross-section Epineurium • Loose areolar tissue with sparse, longitudinally-oriented collagen fibers • Some elastic fibers where epineurium abuts perineurium • Able to accommodate a significant amount of nerve stretching and movement • Increases in thickness where nerves cross joints • Constitutes an increasing proportion of nerves as they increase in size • Epineurial fat helps cushion nerves from compressive injury • Decreased epineurial fat found in patients with diabetes Perineurium • Cellular component composed of laminated fibroblasts of up to 15 layers in thickness which are bounded by a basal lamina • Semi-permeable: inner lamellae have tight junctions, providing a barrier to intercellular transport of macromolecules – Tight junctions can be loosened with topical anaesthetics and with osmotic change Perineurium • Exhibits a slightly positive internal pressure – Fascicular contents herniate upon perineurial injury • Under tension longitudinally – Nerve segment shortens upon transection – may complicate surgical repair as nerve can be stretched only approximately 10% before being inhibited by collagen Endoneurium • Intrafascicular connective tissue consisting of a collagenous matrix in the interstitial space • Develops into partitions of dense connective tissue between diverging fascicles and eventually becomes perineurium when the fascicles separate • Collagen fibers are longitudinally-oriented and run along nerve fibers and capillaries. -

A System for Studying Mechanisms of Neuromuscular Junction Development and Maintenance Valérie Vilmont1,‡, Bruno Cadot1, Gilles Ouanounou2 and Edgar R
© 2016. Published by The Company of Biologists Ltd | Development (2016) 143, 2464-2477 doi:10.1242/dev.130278 TECHNIQUES AND RESOURCES RESEARCH ARTICLE A system for studying mechanisms of neuromuscular junction development and maintenance Valérie Vilmont1,‡, Bruno Cadot1, Gilles Ouanounou2 and Edgar R. Gomes1,3,*,‡ ABSTRACT different animal models and cell lines (Chen et al., 2014; Corti et al., The neuromuscular junction (NMJ), a cellular synapse between a 2012; Lenzi et al., 2015) with the hope of recapitulating some motor neuron and a skeletal muscle fiber, enables the translation of features of neuromuscular diseases and understanding the triggers chemical cues into physical activity. The development of this special of one of their common hallmarks: the disruption of the structure has been subject to numerous investigations, but its neuromuscular junction (NMJ). The NMJ is one of the most complexity renders in vivo studies particularly difficult to perform. studied synapses. It is formed of three key elements: the lower motor In vitro modeling of the neuromuscular junction represents a powerful neuron (the pre-synaptic compartment), the skeletal muscle (the tool to delineate fully the fine tuning of events that lead to subcellular post-synaptic compartment) and the Schwann cell (Sanes and specialization at the pre-synaptic and post-synaptic sites. Here, we Lichtman, 1999). The NMJ is formed in a step-wise manner describe a novel heterologous co-culture in vitro method using rat following a series of cues involving these three cellular components spinal cord explants with dorsal root ganglia and murine primary and its role is basically to ensure the skeletal muscle functionality. -

The Histology of the Neuromuscular Junction In
75 THE HISTOLOGY OF THE NEUROMUSCULAR JUNCTION Downloaded from https://academic.oup.com/brain/article/84/1/75/372729 by guest on 27 September 2021 IN DYSTROPHIA MYOTONICA BY VIOLET MACDERMOT Department of Neurology, St. Thomas' Hospital, London, S.E.I (1) INTRODUCTION DYSTROPJHC MYOTONICA is a familial disease affecting males and females, usually presenting in adult life, characterized by muscular wasting and weakness together with certain other features. The muscles mainly in- volved are the temporal, masseter, facial, sternomastoid and limb muscles, in the latter those mainly affected being peripheral in distribution. A widespread disorder of muscular contraction, myotonia, is also present but is noticed chiefly in the tongue and in the muscles involved in grasping. The other features of the condition are some degree of mental defect, dysphonia, cataracts, frontal baldness, sparse body hair and testicular atrophy. Any of the manifestations of the disease may be absent and the order of presentation of symptoms is variable. The myotonia may precede muscular wasting by many years or may occur independently. In those muscles which are severely wasted the myotonia tends to disappear. The interest of dystrophia myotonica lies in the peculiar distribution of muscle involvement and in the combination of a disorder of muscle function with endocrine and other dysplasic features. The results of histological examination of biopsy and post-mortem material have been described and reviewed by numerous workers, notably Steinert (1909), Adie and Greenfield (1923), Keschner and Davison (1933), Hassin and Kesert (1948), Wohlfart (1951), Adams, Denny-Brown and Pearson (1953), Greenfield, Shy, Alvord and Berg (1957). -

Modulatory Roles of ATP and Adenosine in Cholinergic Neuromuscular Transmission
International Journal of Molecular Sciences Review Modulatory Roles of ATP and Adenosine in Cholinergic Neuromuscular Transmission Ayrat U. Ziganshin 1,* , Adel E. Khairullin 2, Charles H. V. Hoyle 1 and Sergey N. Grishin 3 1 Department of Pharmacology, Kazan State Medical University, 49 Butlerov Street, 420012 Kazan, Russia; [email protected] 2 Department of Biochemistry, Laboratory and Clinical Diagnostics, Kazan State Medical University, 49 Butlerov Street, 420012 Kazan, Russia; [email protected] 3 Department of Medical and Biological Physics with Computer Science and Medical Equipment, Kazan State Medical University, 49 Butlerov Street, 420012 Kazan, Russia; [email protected] * Correspondence: [email protected]; Tel.: +7-843-236-0512 Received: 30 June 2020; Accepted: 1 September 2020; Published: 3 September 2020 Abstract: A review of the data on the modulatory action of adenosine 5’-triphosphate (ATP), the main co-transmitter with acetylcholine, and adenosine, the final ATP metabolite in the synaptic cleft, on neuromuscular transmission is presented. The effects of these endogenous modulators on pre- and post-synaptic processes are discussed. The contribution of purines to the processes of quantal and non- quantal secretion of acetylcholine into the synaptic cleft, as well as the influence of the postsynaptic effects of ATP and adenosine on the functioning of cholinergic receptors, are evaluated. As usual, the P2-receptor-mediated influence is minimal under physiological conditions, but it becomes very important in some pathophysiological situations such as hypothermia, stress, or ischemia. There are some data demonstrating the same in neuromuscular transmission. It is suggested that the role of endogenous purines is primarily to provide a safety factor for the efficiency of cholinergic neuromuscular transmission. -

View Software
TLR4-activated microglia have divergent effects on oligodendrocyte lineage cells DISSERTATION Presented in partial fulfillment of the requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University Evan Zachary Goldstein, BA Neuroscience Graduate Program The Ohio State University 2016 Dissertation Committee: Dr. Dana McTigue, Advisor Dr. Phillip Popovich Dr. Jonathan Godbout Dr. Courtney DeVries Copyright by Evan Zachary Goldstein 2016 i Abstract Myelin accelerates action potential conduction velocity and provides essential metabolic support for axons. Unfortunately, myelin and myelinating cells are often vulnerable to injury or disease, resulting in myelin damage, which in turn can lead to axon dysfunction, overt pathology and neurological impairment. Inflammation is a common component of CNS trauma and disease, and therefore an active inflammatory response is often considered deleterious to myelin health. While inflammation can certainly damage myelin, inflammatory processes also benefit oligodendrocyte (OL) lineage progression and myelin repair. Consistent with the divergent nature of inflammation, intraspinal toll-like receptor 4 (TLR4) activation, an innate immune pathway, kills OL lineage cells, but also initiates oligodendrogenesis. Soluble factors produced by TLR4-activated microglia can reproduce these effects in vitro, however the exact factors are unknown. To determine what microglial factors might contribute to TLR4-induced OL loss and oligodendrogenesis, mRNA of factors known to affect OL lineage cells was quantified in TLR4-activated microglia and spinal cords (chapter 2). Results indicate that TLR4-activated microglia transcribe numerous factors that induce OL loss, OL progenitor cell (OPC) proliferation and OPC differentiation. However, some factors upregulated after intraspinal TLR4 activation were not ii upregulated by microglia, suggesting that other cell types contribute to transcriptional changes in vivo. -
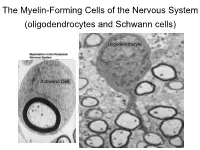
The Myelin-Forming Cells of the Nervous System (Oligodendrocytes and Schwann Cells)
The Myelin-Forming Cells of the Nervous System (oligodendrocytes and Schwann cells) Oligodendrocyte Schwann Cell Oligodendrocyte function Saltatory (jumping) nerve conduction Oligodendroglia PMD PMD Saltatory (jumping) nerve conduction Investigating the Myelinogenic Potential of Individual Oligodendrocytes In Vivo Sparse Labeling of Oligodendrocytes CNPase-GFP Variegated expression under the MBP-enhancer Cerebral Cortex Corpus Callosum Cerebral Cortex Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Corpus Callosum Cerebral Cortex Caudate Putamen Corpus Callosum Ant Commissure Corpus Callosum Cerebral Cortex Caudate Putamen Piriform Cortex Corpus Callosum Ant Commissure Characterization of Oligodendrocyte Morphology Cerebral Cortex Corpus Callosum Caudate Putamen Cerebellum Brain Stem Spinal Cord Oligodendrocytes in disease: Cerebral Palsy ! CP major cause of chronic neurological morbidity and mortality in children ! CP incidence now about 3/1000 live births compared to 1/1000 in 1980 when we started intervening for ELBW ! Of all ELBW {gestation 6 mo, Wt. 0.5kg} , 10-15% develop CP ! Prematurely born children prone to white matter injury {WMI}, principle reason for the increase in incidence of CP ! ! 12 Cerebral Palsy Spectrum of white matter injury ! ! Macro Cystic Micro Cystic Gliotic Khwaja and Volpe 2009 13 Rationale for Repair/Remyelination in Multiple Sclerosis Oligodendrocyte specification oligodendrocytes specified from the pMN after MNs - a ventral source of oligodendrocytes -

Nomina Histologica Veterinaria, First Edition
NOMINA HISTOLOGICA VETERINARIA Submitted by the International Committee on Veterinary Histological Nomenclature (ICVHN) to the World Association of Veterinary Anatomists Published on the website of the World Association of Veterinary Anatomists www.wava-amav.org 2017 CONTENTS Introduction i Principles of term construction in N.H.V. iii Cytologia – Cytology 1 Textus epithelialis – Epithelial tissue 10 Textus connectivus – Connective tissue 13 Sanguis et Lympha – Blood and Lymph 17 Textus muscularis – Muscle tissue 19 Textus nervosus – Nerve tissue 20 Splanchnologia – Viscera 23 Systema digestorium – Digestive system 24 Systema respiratorium – Respiratory system 32 Systema urinarium – Urinary system 35 Organa genitalia masculina – Male genital system 38 Organa genitalia feminina – Female genital system 42 Systema endocrinum – Endocrine system 45 Systema cardiovasculare et lymphaticum [Angiologia] – Cardiovascular and lymphatic system 47 Systema nervosum – Nervous system 52 Receptores sensorii et Organa sensuum – Sensory receptors and Sense organs 58 Integumentum – Integument 64 INTRODUCTION The preparations leading to the publication of the present first edition of the Nomina Histologica Veterinaria has a long history spanning more than 50 years. Under the auspices of the World Association of Veterinary Anatomists (W.A.V.A.), the International Committee on Veterinary Anatomical Nomenclature (I.C.V.A.N.) appointed in Giessen, 1965, a Subcommittee on Histology and Embryology which started a working relation with the Subcommittee on Histology of the former International Anatomical Nomenclature Committee. In Mexico City, 1971, this Subcommittee presented a document entitled Nomina Histologica Veterinaria: A Working Draft as a basis for the continued work of the newly-appointed Subcommittee on Histological Nomenclature. This resulted in the editing of the Nomina Histologica Veterinaria: A Working Draft II (Toulouse, 1974), followed by preparations for publication of a Nomina Histologica Veterinaria.