PDF Output of CLIC (Clustering by Inferred Co-Expression)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

CD8 T Cell Memory Transition and Maintenance of Functional Foxo1
FoxO1 Controls Effector-to-Memory Transition and Maintenance of Functional CD8 T Cell Memory This information is current as Melba Marie Tejera, Eui Ho Kim, Jeremy A. Sullivan, Erin of October 1, 2021. H. Plisch and M. Suresh J Immunol 2013; 191:187-199; Prepublished online 3 June 2013; doi: 10.4049/jimmunol.1300331 http://www.jimmunol.org/content/191/1/187 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2013/06/03/jimmunol.130033 Material 1.DC1 http://www.jimmunol.org/ References This article cites 52 articles, 7 of which you can access for free at: http://www.jimmunol.org/content/191/1/187.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 1, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology FoxO1 Controls Effector-to-Memory Transition and Maintenance of Functional CD8 T Cell Memory Melba Marie Tejera, Eui Ho Kim, Jeremy A. -

1 AGING Supplementary Table 2
SUPPLEMENTARY TABLES Supplementary Table 1. Details of the eight domain chains of KIAA0101. Serial IDENTITY MAX IN COMP- INTERFACE ID POSITION RESOLUTION EXPERIMENT TYPE number START STOP SCORE IDENTITY LEX WITH CAVITY A 4D2G_D 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ B 4D2G_E 52 - 69 52 69 100 100 2.65 Å PCNA X-RAY DIFFRACTION √ C 6EHT_D 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ D 6EHT_E 52 - 71 52 71 100 100 3.2Å PCNA X-RAY DIFFRACTION √ E 6GWS_D 41-72 41 72 100 100 3.2Å PCNA X-RAY DIFFRACTION √ F 6GWS_E 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ G 6GWS_F 41-72 41 72 100 100 2.9Å PCNA X-RAY DIFFRACTION √ H 6IIW_B 2-11 2 11 100 100 1.699Å UHRF1 X-RAY DIFFRACTION √ www.aging-us.com 1 AGING Supplementary Table 2. Significantly enriched gene ontology (GO) annotations (cellular components) of KIAA0101 in lung adenocarcinoma (LinkedOmics). Leading Description FDR Leading Edge Gene EdgeNum RAD51, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, CENPW, HJURP, NDC80, CDCA5, NCAPH, BUB1, ZWILCH, CENPK, KIF2C, AURKA, CENPN, TOP2A, CENPM, PLK1, ERCC6L, CDT1, CHEK1, SPAG5, CENPH, condensed 66 0 SPC24, NUP37, BLM, CENPE, BUB3, CDK2, FANCD2, CENPO, CENPF, BRCA1, DSN1, chromosome MKI67, NCAPG2, H2AFX, HMGB2, SUV39H1, CBX3, TUBG1, KNTC1, PPP1CC, SMC2, BANF1, NCAPD2, SKA2, NUP107, BRCA2, NUP85, ITGB3BP, SYCE2, TOPBP1, DMC1, SMC4, INCENP. RAD51, OIP5, CDK1, SPC25, CCNB1, BIRC5, NCAPG, ZWINT, MAD2L1, SKA3, NUF2, BUB1B, CENPA, SKA1, AURKB, NEK2, ESCO2, CENPW, HJURP, TTK, NDC80, CDCA5, BUB1, ZWILCH, CENPK, KIF2C, AURKA, DSCC1, CENPN, CDCA8, CENPM, PLK1, MCM6, ERCC6L, CDT1, HELLS, CHEK1, SPAG5, CENPH, PCNA, SPC24, CENPI, NUP37, FEN1, chromosomal 94 0 CENPL, BLM, KIF18A, CENPE, MCM4, BUB3, SUV39H2, MCM2, CDK2, PIF1, DNA2, region CENPO, CENPF, CHEK2, DSN1, H2AFX, MCM7, SUV39H1, MTBP, CBX3, RECQL4, KNTC1, PPP1CC, CENPP, CENPQ, PTGES3, NCAPD2, DYNLL1, SKA2, HAT1, NUP107, MCM5, MCM3, MSH2, BRCA2, NUP85, SSB, ITGB3BP, DMC1, INCENP, THOC3, XPO1, APEX1, XRCC5, KIF22, DCLRE1A, SEH1L, XRCC3, NSMCE2, RAD21. -

MRPS12 Rabbit Pab
Leader in Biomolecular Solutions for Life Science MRPS12 Rabbit pAb Catalog No.: A10573 Basic Information Background Catalog No. Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in A10573 protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% Observed MW protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is 14kDa reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins Calculated MW comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical 15kDa properties, which prevents easy recognition by sequence homology. This gene encodes a 28S subunit protein that belongs to the ribosomal protein S12P family. The encoded Category protein is a key component of the ribosomal small subunit and controls the decoding fidelity and susceptibility to aminoglycoside antibiotics. The gene for mitochondrial Primary antibody seryl-tRNA synthetase is located upstream and adjacent to this gene, and both genes are possible candidates for the autosomal dominant deafness gene (DFNA4). Splice Applications variants that differ in the 5' UTR have been found for this gene; all three variants encode WB,IHC the same protein. Cross-Reactivity Human Recommended Dilutions Immunogen Information WB 1:1000 - 1:2000 Gene ID Swiss Prot 6183 O15235 IHC 1:50 - 1:200 Immunogen Recombinant fusion protein containing a sequence corresponding to amino acids 30-138 of human MRPS12 (NP_203527.1). Synonyms MRPS12;MPR-S12;MT-RPS12;RPMS12;RPS12;RPSM12 Contact Product Information www.abclonal.com Source Isotype Purification Rabbit IgG Affinity purification Storage Store at -20℃. -
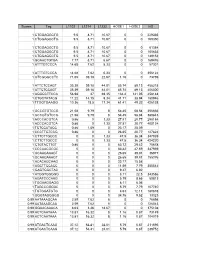
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1
Scores Tag L1102 L1214 L1232 HOSE1 HOSE2 HS 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 229335 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 169350 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 61384 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 105633 1 CTGGAGGCTG 9.5 8.71 10.67 0 0 149152 1 GCAACTGTGA 7.77 8.71 6.67 0 0 169476 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 57301 1 ATTTGTCCCA 14.68 7.62 5.33 0 0 356122 1 GTCGGGCCTC 71.65 39.18 22.67 1.16 0 73769 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 458218 1 ATTCTCCAGT 35.39 39.18 44.01 85.74 89.13 406300 1 AGGGCTTCCA 56.98 37 69.35 134.4 141.05 458148 1 CTGCTATACG 11.22 14.15 9.34 41.71 38.94 180946 1 TTGGTGAAGG 10.36 18.5 17.34 61.41 49.32 426138 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 356666 1 GCCGTGTCCG 21.58 9.79 8 54.45 58.84 380843 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 298184 1 ACCCACGTCA 0.86 0 1.33 27.81 20.77 400124 1 TCTCCATACC 0.86 1.09 0 23.17 25.09 1 CCCTTGTCCG 0.86 0 0 26.65 20.77 127824 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 347939 1 CTTCTTGCCC 0 0 1.33 47.5 36.34 424220 1 CTGTACTTGT 0.86 0 0 63.72 29.42 75678 1 CCCAACGCGC 0 0 0 83.42 47.59 347939 1 GCAAGAAAGT 0 0 0 26.65 39.81 36977 1 GCAAGAAAGT 0 0 0 26.65 39.81 155376 1 ACACAGCAAG 0 0 0 23.17 15.58 1 AGCTTCCACC 0 0 0 11.59 7.79 355542 1 GAGTGGCTAC 0 0 0 9.27 6.92 1 ATGGTGGGGG 0 0 0 8.11 22.5 343586 1 AGATCCCAAG 0 0 0 5.79 8.65 50813 1 TGGAAGGAGG 0 0 0 8.11 6.06 1 TAGCCGGGAC 0 0 0 5.79 7.79 107740 1 TGTGGATGTG 0 0 0 4.63 12.11 180878 1 GGGTAGGGGG 0 0 0 34.76 9.52 13323 0.99 AATAAAGCAA 2.59 7.62 8 0 0 76698 0.99 AATAAAGCAA 2.59 7.62 8 0 0 126043 0.99 GGAACAAACA 8.63 3.26 18.67 0 0 375108 -

Duodenal Mucosal Mitochondrial Gene Expression Is Associated with Delayed Gastric Emptying in Diabetic Gastroenteropathy
Duodenal mucosal mitochondrial gene expression is associated with delayed gastric emptying in diabetic gastroenteropathy Susrutha Puthanmadhom Narayanan, … , Tamas Ordog, Adil E. Bharucha JCI Insight. 2021;6(2):e143596. https://doi.org/10.1172/jci.insight.143596. Research Article Endocrinology Gastroenterology Graphical abstract Find the latest version: https://jci.me/143596/pdf RESEARCH ARTICLE Duodenal mucosal mitochondrial gene expression is associated with delayed gastric emptying in diabetic gastroenteropathy Susrutha Puthanmadhom Narayanan,1 Daniel O’Brien,2 Mayank Sharma,1 Karl Miller,3 Peter Adams,3 João F. Passos,4 Alfonso Eirin,5 Tamas Ordog,4 and Adil E. Bharucha1 1Division of Gastroenterology and Hepatology, Department of Medicine, Mayo Clinic, Rochester, Minnesota, USA. 2Department of Biomedical Statistics and Informatics, Mayo Clinic, Rochester, Minnesota, USA. 3Sanford Burnham Prebys Medical Discovery Institute, San Diego, California, USA. 4Department of Physiology and Biomedical Engineering and 5Division of Nephrology & Hypertension Research, Department of Medicine, Mayo Clinic, Rochester, Minnesota, USA. Hindered by a limited understanding of the mechanisms responsible for diabetic gastroenteropathy (DGE), management is symptomatic. We investigated the duodenal mucosal expression of protein- coding genes and microRNAs (miRNA) in DGE and related them to clinical features. The diabetic phenotype, gastric emptying, mRNA, and miRNA expression and ultrastructure of duodenal mucosal biopsies were compared in 39 DGE patients and 21 controls. Among 3175 differentially expressed genes (FDR < 0.05), several mitochondrial DNA–encoded (mtDNA-encoded) genes (12 of 13 protein coding genes involved in oxidative phosphorylation [OXPHOS], both rRNAs and 9 of 22 transfer RNAs) were downregulated; conversely, nuclear DNA–encoded (nDNA-encoded) mitochondrial genes (OXPHOS) were upregulated in DGE. -

Supplementary Dataset S2
mitochondrial translational termination MRPL28 MRPS26 6 MRPS21 PTCD3 MTRF1L 4 MRPL50 MRPS18A MRPS17 2 MRPL20 MRPL52 0 MRPL17 MRPS33 MRPS15 −2 MRPL45 MRPL30 MRPS27 AURKAIP1 MRPL18 MRPL3 MRPS6 MRPS18B MRPL41 MRPS2 MRPL34 GADD45GIP1 ERAL1 MRPL37 MRPS10 MRPL42 MRPL19 MRPS35 MRPL9 MRPL24 MRPS5 MRPL44 MRPS23 MRPS25 ITB ITB ITB ITB ICa ICr ITL original ICr ICa ITL ICa ITL original ICr ITL ICr ICa mitochondrial translational elongation MRPL28 MRPS26 6 MRPS21 PTCD3 MRPS18A 4 MRPS17 MRPL20 2 MRPS15 MRPL45 MRPL52 0 MRPS33 MRPL30 −2 MRPS27 AURKAIP1 MRPS10 MRPL42 MRPL19 MRPL18 MRPL3 MRPS6 MRPL24 MRPS35 MRPL9 MRPS18B MRPL41 MRPS2 MRPL34 MRPS5 MRPL44 MRPS23 MRPS25 MRPL50 MRPL17 GADD45GIP1 ERAL1 MRPL37 ITB ITB ITB ITB ICa ICr original ICr ITL ICa ITL ICa ITL original ICr ITL ICr ICa translational termination MRPL28 MRPS26 6 MRPS21 PTCD3 C12orf65 4 MTRF1L MRPL50 MRPS18A 2 MRPS17 MRPL20 0 MRPL52 MRPL17 MRPS33 −2 MRPS15 MRPL45 MRPL30 MRPS27 AURKAIP1 MRPL18 MRPL3 MRPS6 MRPS18B MRPL41 MRPS2 MRPL34 GADD45GIP1 ERAL1 MRPL37 MRPS10 MRPL42 MRPL19 MRPS35 MRPL9 MRPL24 MRPS5 MRPL44 MRPS23 MRPS25 ITB ITB ITB ITB ICa ICr original ICr ITL ICa ITL ICa ITL original ICr ITL ICr ICa translational elongation DIO2 MRPS18B MRPL41 6 MRPS2 MRPL34 GADD45GIP1 4 ERAL1 MRPL37 2 MRPS10 MRPL42 MRPL19 0 MRPL30 MRPS27 AURKAIP1 −2 MRPL18 MRPL3 MRPS6 MRPS35 MRPL9 EEF2K MRPL50 MRPS5 MRPL44 MRPS23 MRPS25 MRPL24 MRPS33 MRPL52 EIF5A2 MRPL17 SECISBP2 MRPS15 MRPL45 MRPS18A MRPS17 MRPL20 MRPL28 MRPS26 MRPS21 PTCD3 ITB ITB ITB ITB ICa ICr ICr ITL original ITL ICa ICa ITL ICr ICr ICa original -

Transcriptomic and Proteomic Landscape of Mitochondrial
TOOLS AND RESOURCES Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals Inge Ku¨ hl1,2†*, Maria Miranda1†, Ilian Atanassov3, Irina Kuznetsova4,5, Yvonne Hinze3, Arnaud Mourier6, Aleksandra Filipovska4,5, Nils-Go¨ ran Larsson1,7* 1Department of Mitochondrial Biology, Max Planck Institute for Biology of Ageing, Cologne, Germany; 2Department of Cell Biology, Institute of Integrative Biology of the Cell (I2BC) UMR9198, CEA, CNRS, Univ. Paris-Sud, Universite´ Paris-Saclay, Gif- sur-Yvette, France; 3Proteomics Core Facility, Max Planck Institute for Biology of Ageing, Cologne, Germany; 4Harry Perkins Institute of Medical Research, The University of Western Australia, Nedlands, Australia; 5School of Molecular Sciences, The University of Western Australia, Crawley, Australia; 6The Centre National de la Recherche Scientifique, Institut de Biochimie et Ge´ne´tique Cellulaires, Universite´ de Bordeaux, Bordeaux, France; 7Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden Abstract Dysfunction of the oxidative phosphorylation (OXPHOS) system is a major cause of human disease and the cellular consequences are highly complex. Here, we present comparative *For correspondence: analyses of mitochondrial proteomes, cellular transcriptomes and targeted metabolomics of five [email protected] knockout mouse strains deficient in essential factors required for mitochondrial DNA gene (IKu¨ ); expression, leading to OXPHOS dysfunction. Moreover, -

Anti-MRPL20 Antibody
D225249 Anti-MRPL20 antibody Cat. No. D225249 Package 25 μl/100 μl/200 μl Storage -20°C, pH7.4 PBS, 0.05% NaN3, 40% Glycerol Product overview Description A n ti-MRPL20 rabbit polyclonal antibody Applications ELISA, WB, IHC Immunogen Fusion protein of human MRPL20 Reactivity Human, Mouse Content 1 mg/ml Host species Rabbit Ig class Immunogen-specific rabbit IgG Purification Antigen affinity purification Target information Symbol MRPL20 Full name mitochondrial ribosomal protein L20 Synonyms L20mt; MRP-L20 Swissprot Q9BYC9 Target Background Mammalian mitochondrial ribosomal proteins are encoded by nuclear genes and help in protein synthesis within the mitochondrion. Mitochondrial ribosomes (mitoribosomes) consist of a small 28S subunit and a large 39S subunit. They have an estimated 75% protein to rRNA composition compared to prokaryotic ribosomes, where this ratio is reversed. Another difference between mammalian mitoribosomes and prokaryotic ribosomes is that the latter contain a 5S rRNA. Among different species, the proteins comprising the mitoribosome differ greatly in sequence, and sometimes in biochemical properties, which prevents easy recognition by sequence homology. This gene encodes a 39S subunit protein. A pseudogene corresponding to this gene is found on chromosome 21q. Alternative splicing results in multiple transcript variantsS encodingangon different isoforms. Biotech Applications Immunohistochemistry Predicted cell location: Cytoplasm Predicted cell location: Cytoplasm Positive control: Human liver cancer Positive control: Human brain Recommended dilution: 50-200 Recommended dilution: 50-200 The image on the left is immunohistochemistry of paraffin-embedded The image on the left is immunohistochemistry of paraffin-embedded Human liver cancer tissue using D225249(MRPL20 Antibody) at Human brain tissue using D225249(MRPL20 Antibody) at dilution dilution 1/60, on the right is treated with fusion protein. -

MRPS17 Promotes Invasion and Metastasis Through PI3K/AKT Signal
Journal of Cancer 2021, Vol. 12 4849 Ivyspring International Publisher Journal of Cancer 2021; 12(16): 4849-4861. doi: 10.7150/jca.55719 Research Paper MRPS17 promotes invasion and metastasis through PI3K/AKT signal pathway and could be potential prognostic marker for gastric cancer Wenjie Zhou1,2*, Jun Ouyang1,2*, Junqing Li1,2*, Fangjie Liu4, Tailai An1, Lvjia Cheng1,2, Zi Chong Kuo1, Changhua Zhang1 and Yulong He1,2 1. Digestive Disease Center, The Seventh Affiliated Hospital, Sun Yat-sen University, Zhenyuan Road 628, Guangming District, Shenzhen 518000, Guangdong, China. 2. Department of Gastrointestinal Surgery, The First Affiliated Hospital, Sun Yat-sen University, Zhongshan 2nd Road 58, Yuexiu District, Guangzhou 510080, Guangdong, China. 3. Department of Pathology, The Seventh Affiliated Hospital, Sun Yat-sen University, Zhenyuan Road 628, Guangming District, Shenzhen 518000, Guangdong, China. 4. Department of Hematology, The Seventh Affiliated Hospital, Sun Yat-sen University, 58 Zhongshan 2nd Road 58, Yuexiu District, Guangzhou 510080, Guangdong, China. 5. Scientific Research Centre, The Seventh Affiliated Hospital, Sun Yat-sen University, 628 Zhenyuan Road 628, Guangming District, Shenzhen 518000, Guangdong, China. *These authors contributed equally to this work. Corresponding authors: Changhua Zhang, E-mail: [email protected]; Yulong He, E-mail: [email protected]. © The author(s). This is an open access article distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/4.0/). See http://ivyspring.com/terms for full terms and conditions. Received: 2020.11.10; Accepted: 2021.05.29; Published: 2021.06.11 Abstract In this study, the molecular mechanisms through which Mitochondrial Ribosomal Protein S17 (MRPS17) contributes to gastric cancer (GC) and its prognostic significance in GC have been explored. -

Table SI. List of Differentially Expressed Genes Detected from Three Expression Profiling Datasets GSE18842, GSE30219 and GSE33532
Table SI. List of differentially expressed genes detected from three expression profiling datasets GSE18842, GSE30219 and GSE33532. DEGs Gene Upregulated (n=428) ABCB6, ACP1, ADAM12, ADAMDEC1, ADAMTS12, AFAP1-AS1, AKR1B10, ALDH3B2, ALYREF, ANKRD22, APOBEC3B, ARG2, ARNTL2, ARTN, ASF1B, ATAD2, ATP11B, AUNIP, AURKB, BAIAP2L1, BCL11A, BIK, BIRC5, BLM, BNIP3, BORA, BUB1, BYSL, C10orf2, C12orf56, C12orf66, C15orf48, C2CD4A, C4orf46, C5orf46, CABYR, CALU, CASC7, CBX2, CCNA2, CCNE1, CCNE2, CCT2, CCT6A, CDC25A, CDC25C, CDC45, CDCA2, CDCA3, CDCA8, CDH3, CDK1, CDK5R1, CDKN2A, CDT1, CEMIP, CENPF, CENPH, CENPI, CENPK, CENPL, CENPM, CENPN, MLF1IP, CENPW, CERS6, CHAC2, CHEK1, CIART, CKAP2, CKAP2L, CKAP4, CKS1B, CNFN, COCH, COL11A1, COL3A1, COL5A1, COL5A2, CPD, CRABP2, CST1, CTSV, CTTN, CXCL13, DARS2, DBF4, DCAF13, DCUN1D1, DCUN1D5, DENND1A, DEPDC1, DEPDC1B, DEPDC7, DHFR, DIO2, DONSON, DPY19L1, DSC2, DSCC1, DSG2, DST, DTL, DUS4L, E2F7, ECT2, EGLN3, EIF4EBP1, ELOVL6, ENAH, ENO1, EPT1, ERCC6L, ERO1A, ESPL1, ESRP1, ETV4, EZH2, F12, FAM169A, FAM210A, FAM64A, FAM69A, FAM83A, FAM83D, FAM83F, FAM83H, FANCD2, FANCI, FAP, FAXC, FBXO32, FBXO45, FBXO5, FERMT1, FGF11, FHL2, FLAD1, FNDC1, FOXM1, GAL, GALNT14, GALNT2, GALNT7, GART, GCLC, GCLM, GGH, GINS3, GJB2, GJB6, GMPS, GOLM1, GPX2, GRHL1, GRTP1, GTSE1, H2AFX, HELLS, HILPDA, HIST1H2AE, HIST1H2BC, HIST1H2BH, HIST1H2BJ, HIST1H3F, HIST3H2A, HLTF, HMGA1, HMGA2, HMGB3, HOOK1, HORMAD1, HOXA10, HOXC10, HOXD10, HRASLS, HS6ST2, HSPA4L, HYLS1, IDH2, IGF2BP3, IGFBP2, IGFBP3, IGHM, IGSF9, IL4I1, IQGAP3, IRF6, ITGA11, KCNK1, -
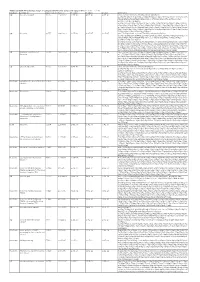
Additional Tables.Xlsx
Additional Table 5 Enriched pathways of upregulated DEGs after spinal cord injury in WT (P < 0.05, q < 0.05) Database Description Ratio of DEGs Ratio of P -value P adjust q value gene name GO ribosomal subunit 87/755 214/12262 1.03E-49 4.61E-47 4.20E-47 Gm6576/Gm10036/Gm5786/Mrps28/Gm9493/Rpl13- ps3/Rpl7l1/Mrpl10/Mrpl44/Rps20/Mrpl51/Mrps11/Mpv17l2/Mrps7/Gm10020/Gm10073 /Mrpl2/Mrpl52/Rpl22/Mrpl4/Mrps24/Zcchc17/Rpl28/Mrpl34/Rpl29/Mrps36/Rpl9- ps6/Hba-a2/Hba-a1/Rpl23a- ps3/Rpl15/Rack1/Mrps18a/Mrpl28/Rpl10a/Mrpl12/Rpl18/Rpl35a/Mrpl41/Mrps12/Rpl36 a/Rps9/Rps18/Rps5/Rps27a/Rps24/Rplp2/Rpl14/Mrpl30/Rps6/Rpl6/Rps3/Rps4x/Rpsa/R pl7/Rpl27a/Rps23/Rps10/Mrps21/Rpl36/Rpl30/Rps17/Mrpl42/Rpl35/Rpl27/Rplp0/Rps28 /Rpl31/Rpl39/Rps13/Rpl13/Rps11/Rpl8/Rps3a1/Fau/Rpl9/Rpl3/Rpl26/Rps15/Rpl13a/Uba 52/Rpl24/Rpl19/Rps14/Rpl4/Rps27/Rplp1 GO ribosome 93/755 248/12262 1.67E-49 4.61E-47 4.20E-47 Gm6576/Pnpt1/Gm10036/Gm5786/Mrps28/Gm9493/Rpl13- ps3/Rpl7l1/Mrpl10/Mrpl44/Rps20/Mrpl51/Mrps11/Mpv17l2/Mrps7/Gm10020/Gm10073 /Mrpl2/Mrpl52/Rpl22/Mrpl4/Mrps24/Zcchc17/Rpl28/Mrpl34/Rpl29/Mrps36/Rpl9- ps6/Hba-a2/Hba-a1/Rpl23a- ps3/Rpl15/Rack1/Eif3h/Mrps18a/Mrpl28/Mrps33/Rpl10a/Mrpl12/Rpl18/Rpl35a/Btf3/Mrp l41/Mrps12/Rpl36a/Rps9/Rps18/Rps5/Rps27a/Rps24/Rplp2/Rpl14/Mrpl30/Rps6/Rpl6/R ps3/Rps4x/Rpsa/Rpl7/Rpl27a/Rps23/Rps10/Mrps21/Rpl36/Rpl30/Rps17/Mrpl42/Rpl35/R pl27/Rplp0/Rps28/Rpl31/Rpl39/Rps13/Rpl13/Rps11/Rpl8/Rps3a1/Fau/Rpl9/Rpl3/Rpl26/ Rps15/Rpl13a/Uba52/Rpl41/Rpl24/Rpl19/Rps14/Rpl4/Ndufa7/Rps27/Rplp1 GO structural constituent of 76/733 169/11958 1.65E-47 1.16E-44 1.12E-44 Gm6576/Gm10036/Gm5786/Rpl7l1/Mrpl10/Rps20/Mrpl51/Mrps11/Mrps7/Gm10020/G -

MRPS12 (NM 033362) Human Tagged ORF Clone – RC202911
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC202911 MRPS12 (NM_033362) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: MRPS12 (NM_033362) Human Tagged ORF Clone Tag: Myc-DDK Symbol: MRPS12 Synonyms: MPR-S12; MT-RPS12; RPMS12; RPS12; RPSM12 Vector: pCMV6-Entry (PS100001) E. coli Selection: Kanamycin (25 ug/mL) Cell Selection: Neomycin ORF Nucleotide >RC202911 ORF sequence Sequence: Red=Cloning site Blue=ORF Green=Tags(s) TTTTGTAATACGACTCACTATAGGGCGGCCGGGAATTCGTCGACTGGATCCGGTACCGAGGAGATCTGCC GCCGCGATCGCC ATGTCCTGGTCTGGCCTTCTCCATGGCCTCAACACGTCCCTAACTTGTGGCCCAGCTCTGGTTCCCCGGC TCTGGGCTACCTGCTCCATGGCTACCCTGAACCAGATGCACCGCCTGGGGCCCCCCAAGCGGCCGCCTCG GAAGCTGGGCCCCACGGAAGGCCGGCCGCAGCTGAAGGGTGTGGTCCTGTGCACGTTTACCCGCAAGCCG AAGAAGCCCAACTCAGCCAATCGCAAGTGCTGTCGAGTGCGGCTCAGCACTGGCCGCGAGGCCGTCTGCT TCATCCCTGGGGAGGGCCACACCCTGCAGGAGCACCAGATTGTCCTTGTGGAGGGCGGCCGCACCCAGGA CCTGCCAGGCGTCAAGCTCACCGTTGTGCGTGGCAAGTACGACTGTGGCCACGTGCAGAAGAAG ACGCGTACGCGGCCGCTCGAGCAGAAACTCATCTCAGAAGAGGATCTGGCAGCAAATGATATCCTGGATT ACAAGGATGACGACGATAAGGTTTAA Protein Sequence: >RC202911 protein sequence Red=Cloning site Green=Tags(s) MSWSGLLHGLNTSLTCGPALVPRLWATCSMATLNQMHRLGPPKRPPRKLGPTEGRPQLKGVVLCTFTRKP KKPNSANRKCCRVRLSTGREAVCFIPGEGHTLQEHQIVLVEGGRTQDLPGVKLTVVRGKYDCGHVQKK TRTRPLEQKLISEEDLAANDILDYKDDDDKV Chromatograms: https://cdn.origene.com/chromatograms/mk6395_c11.zip Restriction Sites: