A-Ailable At
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(12) United States Patent (10) Patent No.: US 6,395,889 B1 Robison (45) Date of Patent: May 28, 2002
USOO6395889B1 (12) United States Patent (10) Patent No.: US 6,395,889 B1 Robison (45) Date of Patent: May 28, 2002 (54) NUCLEIC ACID MOLECULES ENCODING WO WO-98/56804 A1 * 12/1998 ........... CO7H/21/02 HUMAN PROTEASE HOMOLOGS WO WO-99/0785.0 A1 * 2/1999 ... C12N/15/12 WO WO-99/37660 A1 * 7/1999 ........... CO7H/21/04 (75) Inventor: fish E. Robison, Wilmington, MA OTHER PUBLICATIONS Vazquez, F., et al., 1999, “METH-1, a human ortholog of (73) Assignee: Millennium Pharmaceuticals, Inc., ADAMTS-1, and METH-2 are members of a new family of Cambridge, MA (US) proteins with angio-inhibitory activity', The Journal of c: - 0 Biological Chemistry, vol. 274, No. 33, pp. 23349–23357.* (*) Notice: Subject to any disclaimer, the term of this Descriptors of Protease Classes in Prosite and Pfam Data patent is extended or adjusted under 35 bases. U.S.C. 154(b) by 0 days. * cited by examiner (21) Appl. No.: 09/392, 184 Primary Examiner Ponnathapu Achutamurthy (22) Filed: Sep. 9, 1999 ASSistant Examiner William W. Moore (51) Int. Cl." C12N 15/57; C12N 15/12; (74) Attorney, Agent, or Firm-Alston & Bird LLP C12N 9/64; C12N 15/79 (57) ABSTRACT (52) U.S. Cl. .................... 536/23.2; 536/23.5; 435/69.1; 435/252.3; 435/320.1 The invention relates to polynucleotides encoding newly (58) Field of Search ............................... 536,232,235. identified protease homologs. The invention also relates to 435/6, 226, 69.1, 252.3 the proteases. The invention further relates to methods using s s s/ - - -us the protease polypeptides and polynucleotides as a target for (56) References Cited diagnosis and treatment in protease-mediated disorders. -

The Extracellular Proteases Produced by Vibrio Parahaemolyticus
World Journal of Microbiology and Biotechnology (2018) 34:68 https://doi.org/10.1007/s11274-018-2453-4 REVIEW The extracellular proteases produced by Vibrio parahaemolyticus George Osei‑Adjei1 · Xinxiang Huang1 · Yiquan Zhang1 Received: 20 February 2018 / Accepted: 8 May 2018 © Springer Science+Business Media B.V., part of Springer Nature 2018 Abstract Vibrio parahaemolyticus, a Gram-negative bacterium, inhabits marine and estuarine environments and it is a major pathogen responsible globally for most cases of seafood-associated gastroenteritis in humans and acute hepatopancreatic necrosis syn- drome in shrimps. There has been a dramatic worldwide increase in V. parahaemolyticus infections over the last two decades. The pathogenicity of V. parahaemolyticus has been linked to the expression of different kinds of virulence factors including extracellular proteases, such as metalloproteases and serine proteases. V. parahaemolyticus expresses the metalloproteases; PrtV, VppC, VPM and the serine proteases; VPP1/Protease A, VpSP37, PrtA. Extracellular proteases have been identified as potential virulence factors which directly digest many kinds of host proteins or indirectly are involved in the processing of other toxic protein factors. This review summarizes findings on the metalloproteases and serine proteases produced by V. parahaemolyticus and their roles in infections. Identifying the role of V. parahaemolyticus virulence-associated extracellular proteases deepens our understanding of diseases caused by this bacterium. Keywords Extracellular protease · Metalloprotease · Serine protease · Vibrio parahaemolyticus · Virulence Introduction incubation period of V. parahaemolyticus is usually 12–24 h post-infection with infection usually occurring in the hot The Gram-negative bacterium, Vibrio parahaemolyticus, summer months because the bacteria is found free swim- lives in marine and estuarine environments and it is a major ming in coastal water or attached to fish and shellfish. -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -
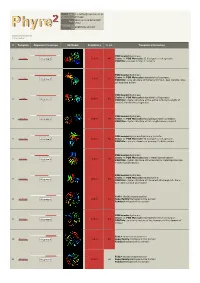
Phyre 2 Results for P22894
Email [email protected] Description P22894 Wed Jun 6 09:54:50 BST Date 2012 Unique Job eba6f50a2e10c91f ID Detailed template information # Template Alignment Coverage 3D Model Confidence % i.d. Template Information PDB header:hydrolase 1 c1gxdA_ Alignment 100.0 46 Chain: A: PDB Molecule:72 kda type iv collagenase; PDBTitle: prommp-2/timp-2 complex PDB header:hydrolase Chain: A: PDB Molecule:interstitial collagenase; 2 c1su3A_ 100.0 61 Alignment PDBTitle: x-ray structure of human prommp-1: new insights into2 collagenase action PDB header:hydrolase Chain: B: PDB Molecule:interstitial collagenase; 3 c2cltB_ 100.0 61 Alignment PDBTitle: crystal structure of the active form (full-length) of human2 fibroblast collagenase. PDB header:hydrolase 4 c3ba0A_ Alignment 100.0 49 Chain: A: PDB Molecule:macrophage metalloelastase; PDBTitle: crystal structure of full-length human mmp-12 PDB header:hydrolase/hydrolase inhibitor 5 c1eakA_ Alignment 100.0 52 Chain: A: PDB Molecule:72 kda type iv collagenase; PDBTitle: catalytic domain of prommp-2 e404q mutant PDB header:hydrolase Chain: A: PDB Molecule:matrix metalloproteinase-9; 6 c1l6jA_ 100.0 56 Alignment PDBTitle: crystal structure of human matrix metalloproteinase mmp92 (gelatinase b). PDB header:hydrolase Chain: A: PDB Molecule:stromelysin-1; 7 c1slmA_ 100.0 54 Alignment PDBTitle: crystal structure of fibroblast stromelysin-1: the c- truncated human2 proenzyme Fold:4-bladed beta-propeller 8 d1fbla1 Alignment 100.0 57 Superfamily:Hemopexin-like domain Family:Hemopexin-like domain PDB header:hydrolase -

Genomic and Proteomic Analyses of the Coral Pathogen Vibrio Coralliilyticus Reveal a Diverse Virulence Repertoire
The ISME Journal (2011) 5, 1471–1483 & 2011 International Society for Microbial Ecology All rights reserved 1751-7362/11 www.nature.com/ismej ORIGINAL ARTICLE Genomic and proteomic analyses of the coral pathogen Vibrio coralliilyticus reveal a diverse virulence repertoire Eidy de O Santos1, Nelson Alves Jr1, Graciela M Dias1, Ana Maria Mazotto2, Alane Vermelho2, Gary J Vora3, Bryan Wilson4, Victor H Beltran4, David G Bourne4, Fre´de´rique Le Roux5,6 and Fabiano L Thompson1 1Laboratory of Microbiology, Institute of Biology, Federal University of Rio de Janeiro (UFRJ), Rio de Janeiro, Brazil; 2Laboratory of Proteases, Institute of Microbiology, UFRJ, Rio de Janeiro, Brazil; 3Center for Bio/ Molecular Science & Engineering, Naval Research Laboratory, Washington, DC, USA; 4Centre for Marine Microbiology and Genetics, Australian Institute of Marine Science, Townsville MC, Queensland, Australia; 5FR2424 CNRS UPMC Station Biologique de Roscoff, France and 6Ifremer, Laboratoire de Physiologie des Inverte´bre´s, Brest, France Vibrio coralliilyticus has been implicated as an important pathogen of coral species worldwide. In this study, the nearly complete genome of Vibrio coralliilyticus strain P1 (LMG23696) was sequenced and proteases implicated in virulence of the strain were specifically investigated. The genome sequence of P1 (5 513 256 bp in size) consisted of 5222 coding sequences and 58 RNA genes (53 tRNAs and at least 5 rRNAs). Seventeen metalloprotease and effector (vgrG, hlyA and hcp) genes were identified in the genome and expressed proteases were also detected in the secretome of P1. As the VcpA zinc-metalloprotease has been considered an important virulence factor of V. coralliilyticus, a vcpA deletion mutant was constructed to evaluate the effect of this gene in animal pathogenesis. -

Composition Containing Protease Produced by Vibrio and Method of Use in Debridement and Wound Healing
Europaisches Patentamt European Patent Office 0 Publication number: 0 472 011 A1 Office europeen des brevets EUROPEAN PATENT APPLICATION 0 Application number: 91112732.2 int. CI 5: A61K 37/48, C12N 9/52, C12N 15/31, //(C12N 15/31, 0 Date of filing: 29.07.91 C12R1:63) 0 Priority: 15.08.90 US 567884 0 Inventor: Fortney, Donald Zane 13.03.91 US 670612 2114 Reese Road Westminster, Md. 21157(US) 0 Date of publication of application: Inventor: Durham, Donald Richard 26.02.92 Bulletin 92/09 20617 Beaver Ridge Road Gaithersburg, Md. 20879(US) 0 Designated Contracting States: AT BE CH DE DK ES FR GB GR IT LI LU NL SE 0 Representative: UEXKULL & STOLBERG 0 Applicant: W.R. Grace & Co.-Conn. Patentanwalte Grace Plaza, 1114 Ave. of the Americas Beselerstrasse 4 New York New York 10036(US) W-2000 Hamburg 52(DE) 0 Composition containing protease produced by vibrio and method of use in debridement and wound healing. 0 A composition for treating wounds comprising at least one pharmaceutically acceptable carrier admixed with an effective amount of a protease selected from the group consisting of: (a) an extracellular neutral protease produced by cultivation of a microorganism belonging to the genus Vibrio, said protease characterized by the following properties: i. hydrolyzes components of necrotic tissue including denatured collagen, elastin and fribrin; ii. does not substantially hydrolyze native tissue in viva, and iii. exhibits stable activity when stored at 25° C in a topical formulation; (b) a protease expressed by recombinant host cells which have been transformed or transfected with an expression vector for said protease (a); and (c) mutants and hybrids of proteases (a) and (b) which are characterized by the properties (i) to (iii). -

Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases
Handbook of Proteolytic Enzymes Second Edition Volume 1 Aspartic and Metallo Peptidases Alan J. Barrett Neil D. Rawlings J. Fred Woessner Editor biographies xxi Contributors xxiii Preface xxxi Introduction ' Abbreviations xxxvii ASPARTIC PEPTIDASES Introduction 1 Aspartic peptidases and their clans 3 2 Catalytic pathway of aspartic peptidases 12 Clan AA Family Al 3 Pepsin A 19 4 Pepsin B 28 5 Chymosin 29 6 Cathepsin E 33 7 Gastricsin 38 8 Cathepsin D 43 9 Napsin A 52 10 Renin 54 11 Mouse submandibular renin 62 12 Memapsin 1 64 13 Memapsin 2 66 14 Plasmepsins 70 15 Plasmepsin II 73 16 Tick heme-binding aspartic proteinase 76 17 Phytepsin 77 18 Nepenthesin 85 19 Saccharopepsin 87 20 Neurosporapepsin 90 21 Acrocylindropepsin 9 1 22 Aspergillopepsin I 92 23 Penicillopepsin 99 24 Endothiapepsin 104 25 Rhizopuspepsin 108 26 Mucorpepsin 11 1 27 Polyporopepsin 113 28 Candidapepsin 115 29 Candiparapsin 120 30 Canditropsin 123 31 Syncephapepsin 125 32 Barrierpepsin 126 33 Yapsin 1 128 34 Yapsin 2 132 35 Yapsin A 133 36 Pregnancy-associated glycoproteins 135 37 Pepsin F 137 38 Rhodotorulapepsin 139 39 Cladosporopepsin 140 40 Pycnoporopepsin 141 Family A2 and others 41 Human immunodeficiency virus 1 retropepsin 144 42 Human immunodeficiency virus 2 retropepsin 154 43 Simian immunodeficiency virus retropepsin 158 44 Equine infectious anemia virus retropepsin 160 45 Rous sarcoma virus retropepsin and avian myeloblastosis virus retropepsin 163 46 Human T-cell leukemia virus type I (HTLV-I) retropepsin 166 47 Bovine leukemia virus retropepsin 169 48 -

Spatiotemporal Regulation of Vibrio Exotoxins by Hlyu and Other Transcriptional Regulators
toxins Review Spatiotemporal Regulation of Vibrio Exotoxins by HlyU and Other Transcriptional Regulators Byoung Sik Kim Department of Food Science and Engineering, ELTEC College of Engineering, Ewha Womans University, Seoul 03760, Korea; [email protected] Received: 8 August 2020; Accepted: 19 August 2020; Published: 22 August 2020 Abstract: After invading a host, bacterial pathogens secrete diverse protein toxins to disrupt host defense systems. To ensure successful infection, however, pathogens must precisely regulate the expression of those exotoxins because uncontrolled toxin production squanders energy. Furthermore, inappropriate toxin secretion can trigger host immune responses that are detrimental to the invading pathogens. Therefore, bacterial pathogens use diverse transcriptional regulators to accurately regulate multiple exotoxin genes based on spatiotemporal conditions. This review covers three major exotoxins in pathogenic Vibrio species and their transcriptional regulation systems. When Vibrio encounters a host, genes encoding cytolysin/hemolysin, multifunctional-autoprocessing repeats-in-toxin (MARTX) toxin, and secreted phospholipases are coordinately regulated by the transcriptional regulator HlyU. At the same time, however, they are distinctly controlled by a variety of other transcriptional regulators. How this coordinated but distinct regulation of exotoxins makes Vibrio species successful pathogens? In addition, anti-virulence strategies that target the coordinating master regulator HlyU and related future research directions are discussed. Keywords: exotoxin; transcriptional regulation; hemolysin; cytolysin; MARTX toxin; secreted phospholipase; HlyU; Vibrio species Key Contribution: During infection, major exotoxin genes in pathogenic Vibrio species are coordinately but distinctly regulated by HlyU and other transcriptional regulators for spatiotemporal expression. 1. Introduction The genus Vibrio is composed of various bacterial species that are metabolically versatile. -

Kinetic and Docking Studies of Inhibitors Targeting the Catalytic Zinc in MA Clan Enzymes
FACULTY OF HEALTH SCIENCE DEPARTMENT OF MEDICAL BIOLOGY TUMOUR BIOLOGY RESEARCH GROUP Kinetic and docking studies of inhibitors targeting the catalytic zinc in MA clan enzymes Stian Sjøli A dissertation for the degree of Philosophiae Doctor October 2011 Table of Contents Acknowledgements…………………………………………………… i List of manuscripts………………………………………………….... iv List of abbreviations………………………………………………….. v Preface………………………………………………………………… vi Aim of Study………………………………………………………….. 1 Introduction …………………………………………………………... 2 1. Enzymes…………………………………………………………... 2 1.1. Kinetics of Catalysis ………………………………………… 3 1.2. Thermodynamics of catalysis ……………………………….. 6 1.3. Breaking peptide bonds with water………………………….. 9 1.4. Binding substrates……………………………………………. 11 1.5. Substrate specificity………………………………………….. 11 1.5.1. Catalytic chamber…………………………………….. 14 1.5.2. Triple helical hydrolysis………………………………. 15 1.6. Families of Metalloprotease………………………………….. 16 1.6.1. Activation mechanism ……………………………….. 17 1.6.2. M4 the Thermolysin family…………………………... 20 Thermolysin………………………………………………. 20 Pseudolysin……………………………………………….. 21 1.6.3. M10A the MMP-1 family…………………………….. 22 Gelatinases ……………………………………………….. 23 MMP-2……………………………………………………. 23 MMP-9……………………………………………………. 24 MMP-14…………………………………………………... 24 1.6.4. M13 the Neprilysin family……………………………. 25 2. Pathological roles for MePs………………………………………. 26 2.1. Tumor metastasis…………………………………………….. 26 2.2. Bacterial invasion and sepsis………………………………… 28 2.3. Extracellular matrix degradation…………………………….. 29 2.4. Blood vessel regulation ……………………………………… -

Peptide Sequence
Peptide Sequence Annotation AADHDG CAS-L1 AAEAISDA M10.005-stromelysin 1 (MMP-3) AAEHDG CAS-L2 AAEYGAEA A01.009-cathepsin D AAGAMFLE M10.007-stromelysin 3 (MMP-11) AAQNASMW A06.001-nodavirus endopeptidase AASGFASP M04.003-vibriolysin ADAHDG CAS-L3 ADAPKGGG M02.006-angiotensin-converting enzyme 2 ADATDG CAS-L5 ADAVMDNP A01.009-cathepsin D ADDPDG CAS-21 ADEPDG CAS-L11 ADETDG CAS-22 ADEVDG CAS-23 ADGKKPSS S01.233-plasmin AEALERMF A01.009-cathepsin D AEEQGVTD C03.007-rhinovirus picornain 3C AETFYVDG A02.001-HIV-1 retropepsin AETWYIDG A02.007-feline immunodeficiency virus retropepsin AFAHDG CAS-L24 AFATDG CAS-25 AFDHDG CAS-L26 AFDTDG CAS-27 AFEHDG CAS-28 AFETDG CAS-29 AFGHDG CAS-30 AFGTDG CAS-31 AFQHDG CAS-32 AFQTDG CAS-33 AFSHDG CAS-L34 AFSTDG CAS-35 AFTHDG CAS-L36 AGERGFFY Insulin B-chain AGLQRGGG M14.004-carboxypeptidase N AGSHLVEA Insulin B-chain AIDIDG CAS-L37 AIDPDG CAS-38 AIDTDG CAS-39 AIDVDG CAS-L40 AIEHDG CAS-L41 AIEIDG CAS-L42 AIENDG CAS-43 AIEPDG CAS-44 AIEQDG CAS-45 AIESDG CAS-46 AIETDG CAS-47 AIEVDG CAS-48 AIFQGPID C03.007-rhinovirus picornain 3C AIGHDG CAS-49 AIGNDG CAS-L50 AIGPDG CAS-L51 AIGQDG CAS-52 AIGSDG CAS-53 AIGTDG CAS-54 AIPMSIPP M10.051-serralysin AISHDG CAS-L55 AISNDG CAS-L56 AISPDG CAS-57 AISQDG CAS-58 AISSDG CAS-59 AISTDG CAS-L60 AKQRAKRD S08.071-furin AKRQGLPV C03.007-rhinovirus picornain 3C AKRRAKRD S08.071-furin AKRRTKRD S08.071-furin ALAALAKK M11.001-gametolysin ALDIDG CAS-L61 ALDPDG CAS-62 ALDTDG CAS-63 ALDVDG CAS-L64 ALEIDG CAS-L65 ALEPDG CAS-L66 ALETDG CAS-67 ALEVDG CAS-68 ALFQGPLQ C03.001-poliovirus-type picornain -

Computational Design and Experimental Characterization of Metallopeptides As Proteases for Bioengineering Applications
Henrique Daniel Figueiredo Carvalho Mestre em Bioquímica Computational design and experimental characterization of metallopeptides as proteases for bioengineering applications Dissertação para obtenção do Grau de Doutor em Bioengenharia (MIT Portugal) Orientador: Olga Iranzo, Doutora., Aix-Marseille Université Co-orientadores: Ana Cecília Roque, Prof. Doutora., Univ. NOVA de Lisboa Ricardo J. F. Branco, Doutor, Univ. NOVA de Lisboa Setembro 2017 ii Computational design and experimental characterization of metallopeptides as proteases for bioengineering applications Copyright © Henrique Daniel Figueiredo Carvalho, Faculdade de Ciências e Tecnologia, Universidade Nova de Lisboa. A Faculdade de Ciências e Tecnologia e a Universidade Nova de Lisboa têm o direito, perpétuo e sem limites geográficos, de arquivar e publicar esta dissertação através de exemplares impressos reproduzidos em papel ou de forma digital, ou por qualquer outro meio conhecido ou que venha a ser inventado, e de a divulgar através de repositórios científicos e de admitir a sua cópia e distribuição com objectivos educacionais ou de investigação, não comerciais, desde que seja dado crédito ao autor e editor. iii iv Agradecimentos Este trabalho marca o final de uma importante etapa no meu percurso académico e pessoal e portanto gostaria de agradecer a todos aqueles que de alguma forma estiveram envolvidos. À minha orientadora Doutora Olga Iranzo e co-orientadores Professora Doutora Ana Cecília A. Roque e Ricardo J. F. Branco por propocionarem as condições necessárias à realização deste trabalho nos grupo de Engenharia Biomolecular (UCIBIO/REQUIMTE, Faculdade de Ciências e Tecnologia da Universidade NOVA de Lisboa) e no grupo BiosCiences (Institut des Sciences Moléculaires de Marseille UMR CNRS 7313, Aix-Marseille Université), pela supervisão e apoio crítico na realização tarefas e discussão de resultados. -

Multiple Architectures and Mechanisms of Latency in Metallopeptidase Zymogens
Arolas et al. 1 Multiple architectures and mechanisms of latency in metallopeptidase zymogens Joan L. Arolas a,1, Theodoros Goulas 1, Anna Cuppari and F. Xavier Gomis-Rüth * Proteolysis Laboratory; Structural Biology Unit ("María-de-Maeztu" Unit of Excellence); Molecular Biology Institute of Barcelona (CSIC); Barcelona Science Park; c/Baldiri Reixac, 15-21; 08028 Barcelona (Catalonia, Spain). a Present address: Max F. Perutz Laboratories; University of Vienna; Campus Vienna Biocenter 5; 1030 Vienna (Austria). * Corresponding author: Tel.:(+34) 934 020 186; E-mail: [email protected]. 1 These authors contributed equally and share first authorship. ABSTRACT Metallopeptidases cleave polypeptides bound in the active-site cleft of catalytic domains through a general base/acid- mechanism. This involves a solvent molecule bound to a catalytic zinc and general regulation of the mechanism through zymogen-based latency. Sixty reported structures from eleven metallopeptidase families reveal that pro-segments, mostly N-terminally of the catalytic domain, block the cleft regardless of their size. Pro-segments may be peptides (5-14 residues), which are only structured within the zymogens, or large moieties (<227 residues) of one or two folded domains. While some pro-segments globally shield the catalytic domain through a few contacts, others specifically run across the cleft in the same or opposite direction of a substrate, making numerous interactions. Some pro-segments block the zinc by replacing the solvent with particular side chains, others use terminal α-amino or carboxylate groups. Overall, metallopeptidase zymogens employ disparate mechanisms that diverge even within families, which supports that latency is less conserved than catalysis. CONTENTS 1.