Plastid Phylogenomic Insights Into the Evolution Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
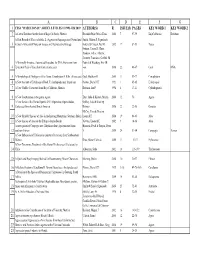
Haseltonia Articles and Authors.Xlsx
ABCDEFG 1 CSSA "HASELTONIA" ARTICLE TITLES #1 1993–#26 2019 AUTHOR(S) R ISSUE(S) PAGES KEY WORD 1 KEY WORD 2 2 A Cactus Database for the State of Baja California, Mexico Resendiz Ruiz, María Elena 2000 7 97-99 BajaCalifornia Database A First Record of Yucca aloifolia L. (Agavaceae/Asparagaceae) Naturalized Smith, Gideon F, Figueiredo, 3 in South Africa with Notes on its uses and Reproductive Biology Estrela & Crouch, Neil R 2012 17 87-93 Yucca Fotinos, Tonya D, Clase, Teodoro, Veloz, Alberto, Jimenez, Francisco, Griffith, M A Minimally Invasive, Automated Procedure for DNA Extraction from Patrick & Wettberg, Eric JB 4 Epidermal Peels of Succulent Cacti (Cactaceae) von 2016 22 46-47 Cacti DNA 5 A Morphological Phylogeny of the Genus Conophytum N.E.Br. (Aizoaceae) Opel, Matthew R 2005 11 53-77 Conophytum 6 A New Account of Echidnopsis Hook. F. (Asclepiadaceae: Stapeliae) Plowes, Darrel CH 1993 1 65-85 Echidnopsis 7 A New Cholla (Cactaceae) from Baja California, Mexico Rebman, Jon P 1998 6 17-21 Cylindropuntia 8 A New Combination in the genus Agave Etter, Julia & Kristen, Martin 2006 12 70 Agave A New Series of the Genus Opuntia Mill. (Opuntieae, Opuntioideae, Oakley, Luis & Kiesling, 9 Cactaceae) from Austral South America Roberto 2016 22 22-30 Opuntia McCoy, Tom & Newton, 10 A New Shrubby Species of Aloe in the Imatong Mountains, Southern Sudan Leonard E 2014 19 64-65 Aloe 11 A New Species of Aloe on the Ethiopia-Sudan Border Newton, Leonard E 2002 9 14-16 Aloe A new species of Ceropegia sect. -

The First Missouri Occurrences of Cerastium Dubium (Anomalous Mouse-Eared Chickweed)
Missouriensis, 34: 20-23. 2017. *pdf effectively published online 30 September 2017 via https://monativeplants.org/missouriensis The first Missouri occurrences of Cerastium dubium (anomalous mouse-eared chickweed) STEVE R. TURNER1 & GERRIT DAVIDSE2 ABSTRACT. – Cerastium dubium is reported new to the Missouri flora from two counties in eastern Missouri. A detailed description is provided based on local populations. Cerastium dubium (Bastard) Guépin (= Cerastium anomalum Waldst. & Kit. ex Willd.; Stellaria dubia Bastard; Dichodon viscidum (M. Bieb.) Holub – see Tropicos.org) is a Eurasian member of the Caryophyllaceae. Commonly called anomalous mouse-eared chickweed, three- styled chickweed, or doubtful chickweed, its first reported appearance in North America was in Washington state in 1966 (Hitchcock and Cronquist 1973). Since then, the plant has been discovered in Illinois (Shildneck and Jones 1986), and is now known from scattered populations in several states bordering Missouri: Illinois, Kentucky, Tennessee, Arkansas, and Kansas. Yatskievych (2006) discusses C. dubium and mentions that although the species had not been reported in Missouri, its arrival is anticipated. According to Yatskievych, the plant generally resembles C. nutans or C. brachypodum, but with the unique characters of three styles and a straight capsule with 6 apical teeth at dehiscence. In March of 2012, while rototilling a garden plot at his residence near Labadie, in Franklin County, Missouri, the first author discovered a small population of plants unfamiliar to him, growing with Lamium amplexicaule and Stellaria media. The flowers of this plant were somewhat showier than the common Stellaria, with wider and less deeply cleft petals. The centers of the flowers were bright yellow due to anthers and profusely shed pollen. -

The Vascular Plants of Massachusetts
The Vascular Plants of Massachusetts: The Vascular Plants of Massachusetts: A County Checklist • First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Somers Bruce Sorrie and Paul Connolly, Bryan Cullina, Melissa Dow Revision • First A County Checklist Plants of Massachusetts: Vascular The A County Checklist First Revision Melissa Dow Cullina, Bryan Connolly, Bruce Sorrie and Paul Somers Massachusetts Natural Heritage & Endangered Species Program Massachusetts Division of Fisheries and Wildlife Natural Heritage & Endangered Species Program The Natural Heritage & Endangered Species Program (NHESP), part of the Massachusetts Division of Fisheries and Wildlife, is one of the programs forming the Natural Heritage network. NHESP is responsible for the conservation and protection of hundreds of species that are not hunted, fished, trapped, or commercially harvested in the state. The Program's highest priority is protecting the 176 species of vertebrate and invertebrate animals and 259 species of native plants that are officially listed as Endangered, Threatened or of Special Concern in Massachusetts. Endangered species conservation in Massachusetts depends on you! A major source of funding for the protection of rare and endangered species comes from voluntary donations on state income tax forms. Contributions go to the Natural Heritage & Endangered Species Fund, which provides a portion of the operating budget for the Natural Heritage & Endangered Species Program. NHESP protects rare species through biological inventory, -

A Preliminary Report of the Biology of the Genus Charpentiera (Amaranthaceae) I
Pacific Science (1973), Vol. 27, No.4, p. 399-405 Printed in Great Britain A Preliminary Report of the Biology of the Genus Charpentiera (Amaranthaceae) I S. H. SOHMER 2 ABSTRACT: The genus Charpentiera (Amaranthaceae), found in the Hawaiian and Austral archipelagoes, has a structurally gynodioecious but functionally dioecious breeding system. The sex ratio varies from taxon to taxon within the genus. The sex-determining mechanism is unknown. A high rate ofovule sterility is found in all the taxa, which i~ not predicted by the pollen sterility figures reported. Data concern ing seed size, pollen size, and seed germination potential are provided. Hybridization is demonstrated to occur between Charpentiera densiflora Sohmer and C. elliptica (Hilleb.) Heller. Reproductive isolation is reported for C. tomentosa var. tomentosa Sohmer and C. obovata Gaud. THE GENUS Charpentiera is indigenous to the often small and thinly scattered throughout the Hawaiian Islands. In 1934 it was also found mature ecosystems of the generic range. Popu on two of the Austral Islands approximately lations of several of the taxa, however, are 2,800 miles disjunct from Hawaii (Suessenguth sometimes numerous in certain areas such as 1936). The members of the genus are trees the heads of deep gulches in some of the highly ranging from about 8 to 40 feet high and are dissected volcanic ranges of the islands. During of diverse habit, with pendant, compound the course of the revision, the reproductive panicles of minute, anemophilous flowers. The biology of Charpentiera was investigated. fruit is indehiscent and uniovulate. A recent systematic revision of Charpentiera (Sohmer 1972) has recognized six major taxa in the SEX RATIO AND TENDENCY TO DIOECISM genus. -

Evaluation of a Proposed Significant Natural Area at Mt Iron, Wanaka
EVALUATION OF A PROPOSED SIGNIFICANT NATURAL AREA AT MT IRON, WANAKA R3762 EVALUATION OF A PROPOSED SIGNIFICANT NATURAL AREA AT MT IRON, WANAKA Coprosma shrubland on the southwest faces at the Allenby Farms site, Mt Iron. Contract Report No. 3762 March 2017 (Revised and updated) Project Team: Kelvin Lloyd - Report author: vegetation and flora Mandy Tocher - Report author: herpetofauna Brian Patrick - Report author: invertebrates Prepared for: Allenby Farms Ltd P.O. Box 196 Wanaka 9343 DUNEDIN OFFICE: 764 CUMBERLAND STREET, DUNEDIN 9016 Ph 03-477-2096, 03-477-2095 HEAD OFFICE: 99 SALA STREET, P.O. BOX 7137, TE NGAE, ROTORUA Ph 07-343-9017, 07-343-9018; email [email protected], www.wildlands.co.nz CONTENTS 1. INTRODUCTION 1 2. SITE CONTEXT 1 3. METHODS 1 4. ECOLOGICAL CONTEXT 4 5. INDIGENOUS VEGETATION AND HABITATS 5 5.1 Kānuka scrub and shrubland 5 5.2 Coprosma scrub and shrubland 6 5.3 Exotic grassland and herbfield 7 5.4 Swale turf 8 5.5 Cushionfield 8 6. FLORA 8 6.1 Species richness 8 6.2 Threatened and At Risk plant species 12 6.3 Pest plants 12 7. BIRDS 13 8. LIZARDS 14 8.1 Overview 14 8.2 “Remove from SNA” zone 14 8.3 Alternate SNA 18 9. INVERTEBRATES 18 9.1 Overview 18 9.2 Mixed Coprosma-dominant shrubland 18 9.3 Kānuka scrub and shrubland 19 9.4 Rock outcrop habitats 19 9.5 Open grassland and turf 19 10. PEST ANIMALS 20 11. ECOLOGICAL VALUES 20 11.1 District Plan (2009) - Section 6c Significance 20 11.2 Proposed District Plan - Section 6c Significance from Policy 33.2.1.9 22 11.3 Significance summary 23 12. -

Urera Kaalae
Plants Opuhe Urera kaalae SPECIES STATUS: Federally Listed as Endangered Genetic Safety Net Species J.K.Obata©Smithsonian Inst., 2005 IUCN Red List Ranking – Critically Endangered (CR D) Hawai‘i Natural Heritage Ranking ‐ Critically Imperiled (G1) Endemism – O‘ahu Critical Habitat ‐ Designated SPECIES INFORMATION: Urera kaalae, a long‐lived perennial member of the nettle family (Urticaceae), is a small tree or shrub 3 to 7 m (10 to 23 ft) tall. This species can be distinguished from the other Hawaiian species of the genus by its heart‐shaped leaves. DISTRIBUTION: Found in the central to southern parts of the Wai‘anae Mountains on O‘ahu. ABUNDANCE: The nine remaining subpopulations comprise approximately 40 plants. LOCATION AND CONDITION OF KEY HABITAT: Urera kaalae typically grows on slopes and in gulches in diverse mesic forest at elevations of 439 to 1,074 m (1,440 to 3,523 ft). The last 12 known occurrences are found on both state and privately owned land. Associated native species include Alyxia oliviformis, Antidesma platyphyllum, Asplenium kaulfusii, Athyrium sp., Canavalia sp., Charpentiera sp., Chamaesyce sp., Claoxylon sandwicense, Diospyros hillebrandii, Doryopteris sp., Freycinetia arborea, Hedyotis acuminata, Hibiscus sp., Nestegis sandwicensis, Pipturus albidus, Pleomele sp., Pouteria sandwicensis, Psychotria sp., Senna gaudichaudii (kolomona), Streblus pendulinus, Urera glabra, and Xylosma hawaiiense. THREATS: Habitat degradation by feral pigs; Competition from alien plant species; Stochastic extinction; Reduced reproductive vigor due to the small number of remaining individuals. CONSERVATION ACTIONS: The goals of conservation actions are not only to protect current populations, but also to establish new populations to reduce the risk of extinction. -

Outline of Angiosperm Phylogeny
Outline of angiosperm phylogeny: orders, families, and representative genera with emphasis on Oregon native plants Priscilla Spears December 2013 The following listing gives an introduction to the phylogenetic classification of the flowering plants that has emerged in recent decades, and which is based on nucleic acid sequences as well as morphological and developmental data. This listing emphasizes temperate families of the Northern Hemisphere and is meant as an overview with examples of Oregon native plants. It includes many exotic genera that are grown in Oregon as ornamentals plus other plants of interest worldwide. The genera that are Oregon natives are printed in a blue font. Genera that are exotics are shown in black, however genera in blue may also contain non-native species. Names separated by a slash are alternatives or else the nomenclature is in flux. When several genera have the same common name, the names are separated by commas. The order of the family names is from the linear listing of families in the APG III report. For further information, see the references on the last page. Basal Angiosperms (ANITA grade) Amborellales Amborellaceae, sole family, the earliest branch of flowering plants, a shrub native to New Caledonia – Amborella Nymphaeales Hydatellaceae – aquatics from Australasia, previously classified as a grass Cabombaceae (water shield – Brasenia, fanwort – Cabomba) Nymphaeaceae (water lilies – Nymphaea; pond lilies – Nuphar) Austrobaileyales Schisandraceae (wild sarsaparilla, star vine – Schisandra; Japanese -

State of New York City's Plants 2018
STATE OF NEW YORK CITY’S PLANTS 2018 Daniel Atha & Brian Boom © 2018 The New York Botanical Garden All rights reserved ISBN 978-0-89327-955-4 Center for Conservation Strategy The New York Botanical Garden 2900 Southern Boulevard Bronx, NY 10458 All photos NYBG staff Citation: Atha, D. and B. Boom. 2018. State of New York City’s Plants 2018. Center for Conservation Strategy. The New York Botanical Garden, Bronx, NY. 132 pp. STATE OF NEW YORK CITY’S PLANTS 2018 4 EXECUTIVE SUMMARY 6 INTRODUCTION 10 DOCUMENTING THE CITY’S PLANTS 10 The Flora of New York City 11 Rare Species 14 Focus on Specific Area 16 Botanical Spectacle: Summer Snow 18 CITIZEN SCIENCE 20 THREATS TO THE CITY’S PLANTS 24 NEW YORK STATE PROHIBITED AND REGULATED INVASIVE SPECIES FOUND IN NEW YORK CITY 26 LOOKING AHEAD 27 CONTRIBUTORS AND ACKNOWLEGMENTS 30 LITERATURE CITED 31 APPENDIX Checklist of the Spontaneous Vascular Plants of New York City 32 Ferns and Fern Allies 35 Gymnosperms 36 Nymphaeales and Magnoliids 37 Monocots 67 Dicots 3 EXECUTIVE SUMMARY This report, State of New York City’s Plants 2018, is the first rankings of rare, threatened, endangered, and extinct species of what is envisioned by the Center for Conservation Strategy known from New York City, and based on this compilation of The New York Botanical Garden as annual updates thirteen percent of the City’s flora is imperiled or extinct in New summarizing the status of the spontaneous plant species of the York City. five boroughs of New York City. This year’s report deals with the City’s vascular plants (ferns and fern allies, gymnosperms, We have begun the process of assessing conservation status and flowering plants), but in the future it is planned to phase in at the local level for all species. -

Species Classification and Nomenclature by Norbert Leist and Andrea Jonitz Prof
ISTA Purity Seminar 15. June 2009 Zürich TlTools for seed identifi cati on species classification and nomenclature by Norbert Leist and Andrea Jonitz Prof. Dr. Norbert Leist Dr. Andrea Jonitz Brahmsstr.25 LTZ Augustenberg 76669 Bad Schönborn Neßlerstr.23 Germany 76227 Karlsruhe [email protected] Germany [email protected] Aquilegia vulgaris, Variation Variation • Variation is everywhere in biological systems. Natural variation at the population level is usualy not continuous, but occurs in discrete units or taxa. Easily the most important taxonomic level is the species because it is often the smallest clearly recognizable and discrete set of populations. • Understanding how species form and how to recognize them have been major challenges to systematists. The variation in one population becomes interrupted, the way to a split into two species strong hairy nearly glabrous Variation on species • Sources of variation: MttiMutation Recombination Independent assortment of the chromosomes Random genetic drift Selection Conservation of species characteristics avoiding gene flow Isolating barriers: temporal (seasonal, diurnal) habitat (wet, dry; calceous, silicious) floral (structural, behavioral eg. adaptations for pollinators) reproductive mode (self fertilisation, agamospery) incompatibility (pollen, seeds) hybrid inviability hybrid floral isolation hybrid sterility hybrid break down Iris germanica Iris sibirica Isolation by habitat Definition of „species“ is not easy A species is the smallest aggregation of populations -

Evolutionary History of Floral Key Innovations in Angiosperms Elisabeth Reyes
Evolutionary history of floral key innovations in angiosperms Elisabeth Reyes To cite this version: Elisabeth Reyes. Evolutionary history of floral key innovations in angiosperms. Botanics. Université Paris Saclay (COmUE), 2016. English. NNT : 2016SACLS489. tel-01443353 HAL Id: tel-01443353 https://tel.archives-ouvertes.fr/tel-01443353 Submitted on 23 Jan 2017 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. NNT : 2016SACLS489 THESE DE DOCTORAT DE L’UNIVERSITE PARIS-SACLAY, préparée à l’Université Paris-Sud ÉCOLE DOCTORALE N° 567 Sciences du Végétal : du Gène à l’Ecosystème Spécialité de Doctorat : Biologie Par Mme Elisabeth Reyes Evolutionary history of floral key innovations in angiosperms Thèse présentée et soutenue à Orsay, le 13 décembre 2016 : Composition du Jury : M. Ronse de Craene, Louis Directeur de recherche aux Jardins Rapporteur Botaniques Royaux d’Édimbourg M. Forest, Félix Directeur de recherche aux Jardins Rapporteur Botaniques Royaux de Kew Mme. Damerval, Catherine Directrice de recherche au Moulon Président du jury M. Lowry, Porter Curateur en chef aux Jardins Examinateur Botaniques du Missouri M. Haevermans, Thomas Maître de conférences au MNHN Examinateur Mme. Nadot, Sophie Professeur à l’Université Paris-Sud Directeur de thèse M. -

Full of Beans: a Study on the Alignment of Two Flowering Plants Classification Systems
Full of beans: a study on the alignment of two flowering plants classification systems Yi-Yun Cheng and Bertram Ludäscher School of Information Sciences, University of Illinois at Urbana-Champaign, USA {yiyunyc2,ludaesch}@illinois.edu Abstract. Advancements in technologies such as DNA analysis have given rise to new ways in organizing organisms in biodiversity classification systems. In this paper, we examine the feasibility of aligning two classification systems for flowering plants using a logic-based, Region Connection Calculus (RCC-5) ap- proach. The older “Cronquist system” (1981) classifies plants using their mor- phological features, while the more recent Angiosperm Phylogeny Group IV (APG IV) (2016) system classifies based on many new methods including ge- nome-level analysis. In our approach, we align pairwise concepts X and Y from two taxonomies using five basic set relations: congruence (X=Y), inclusion (X>Y), inverse inclusion (X<Y), overlap (X><Y), and disjointness (X!Y). With some of the RCC-5 relationships among the Fabaceae family (beans family) and the Sapindaceae family (maple family) uncertain, we anticipate that the merging of the two classification systems will lead to numerous merged solutions, so- called possible worlds. Our research demonstrates how logic-based alignment with ambiguities can lead to multiple merged solutions, which would not have been feasible when aligning taxonomies, classifications, or other knowledge or- ganization systems (KOS) manually. We believe that this work can introduce a novel approach for aligning KOS, where merged possible worlds can serve as a minimum viable product for engaging domain experts in the loop. Keywords: taxonomy alignment, KOS alignment, interoperability 1 Introduction With the advent of large-scale technologies and datasets, it has become increasingly difficult to organize information using a stable unitary classification scheme over time. -

South American Cacti in Time and Space: Studies on the Diversification of the Tribe Cereeae, with Particular Focus on Subtribe Trichocereinae (Cactaceae)
Zurich Open Repository and Archive University of Zurich Main Library Strickhofstrasse 39 CH-8057 Zurich www.zora.uzh.ch Year: 2013 South American Cacti in time and space: studies on the diversification of the tribe Cereeae, with particular focus on subtribe Trichocereinae (Cactaceae) Lendel, Anita Posted at the Zurich Open Repository and Archive, University of Zurich ZORA URL: https://doi.org/10.5167/uzh-93287 Dissertation Published Version Originally published at: Lendel, Anita. South American Cacti in time and space: studies on the diversification of the tribe Cereeae, with particular focus on subtribe Trichocereinae (Cactaceae). 2013, University of Zurich, Faculty of Science. South American Cacti in Time and Space: Studies on the Diversification of the Tribe Cereeae, with Particular Focus on Subtribe Trichocereinae (Cactaceae) _________________________________________________________________________________ Dissertation zur Erlangung der naturwissenschaftlichen Doktorwürde (Dr.sc.nat.) vorgelegt der Mathematisch-naturwissenschaftlichen Fakultät der Universität Zürich von Anita Lendel aus Kroatien Promotionskomitee: Prof. Dr. H. Peter Linder (Vorsitz) PD. Dr. Reto Nyffeler Prof. Dr. Elena Conti Zürich, 2013 Table of Contents Acknowledgments 1 Introduction 3 Chapter 1. Phylogenetics and taxonomy of the tribe Cereeae s.l., with particular focus 15 on the subtribe Trichocereinae (Cactaceae – Cactoideae) Chapter 2. Floral evolution in the South American tribe Cereeae s.l. (Cactaceae: 53 Cactoideae): Pollination syndromes in a comparative phylogenetic context Chapter 3. Contemporaneous and recent radiations of the world’s major succulent 86 plant lineages Chapter 4. Tackling the molecular dating paradox: underestimated pitfalls and best 121 strategies when fossils are scarce Outlook and Future Research 207 Curriculum Vitae 209 Summary 211 Zusammenfassung 213 Acknowledgments I really believe that no one can go through the process of doing a PhD and come out without being changed at a very profound level.