Chem. Pharm. Bull. 51(10) 1217ム1219
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -

Flow Reagents Single Color Antibodies CD Chart
CD CHART CD N° Alternative Name CD N° Alternative Name CD N° Alternative Name Beckman Coulter Clone Beckman Coulter Clone Beckman Coulter Clone T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells T Cells B Cells Granulocytes NK Cells Macrophages/Monocytes Platelets Erythrocytes Stem Cells Dendritic Cells Endothelial Cells Epithelial Cells CD1a T6, R4, HTA1 Act p n n p n n S l CD99 MIC2 gene product, E2 p p p CD223 LAG-3 (Lymphocyte activation gene 3) Act n Act p n CD1b R1 Act p n n p n n S CD99R restricted CD99 p p CD224 GGT (γ-glutamyl transferase) p p p p p p CD1c R7, M241 Act S n n p n n S l CD100 SEMA4D (semaphorin 4D) p Low p p p n n CD225 Leu13, interferon induced transmembrane protein 1 (IFITM1). p p p p p CD1d R3 Act S n n Low n n S Intest CD101 V7, P126 Act n p n p n n p CD226 DNAM-1, PTA-1 Act n Act Act Act n p n CD1e R2 n n n n S CD102 ICAM-2 (intercellular adhesion molecule-2) p p n p Folli p CD227 MUC1, mucin 1, episialin, PUM, PEM, EMA, DF3, H23 Act p CD2 T11; Tp50; sheep red blood cell (SRBC) receptor; LFA-2 p S n p n n l CD103 HML-1 (human mucosal lymphocytes antigen 1), integrin aE chain S n n n n n n n l CD228 Melanotransferrin (MT), p97 p p CD3 T3, CD3 complex p n n n n n n n n n l CD104 integrin b4 chain; TSP-1180 n n n n n n n p p CD229 Ly9, T-lymphocyte surface antigen p p n p n -

The Poliovirus Receptor (CD155)
Cutting Edge: CD96 (Tactile) Promotes NK Cell-Target Cell Adhesion by Interacting with the Poliovirus Receptor (CD155) This information is current as Anja Fuchs, Marina Cella, Emanuele Giurisato, Andrey S. of September 27, 2021. Shaw and Marco Colonna J Immunol 2004; 172:3994-3998; ; doi: 10.4049/jimmunol.172.7.3994 http://www.jimmunol.org/content/172/7/3994 Downloaded from References This article cites 19 articles, 8 of which you can access for free at: http://www.jimmunol.org/content/172/7/3994.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 27, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2004 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. THE JOURNAL OF IMMUNOLOGY CUTTING EDGE Cutting Edge: CD96 (Tactile) Promotes NK Cell-Target Cell Adhesion by Interacting with the Poliovirus Receptor (CD155) Anja Fuchs, Marina Cella, Emanuele Giurisato, Andrey S. Shaw, and Marco Colonna1 The poliovirus receptor (PVR) belongs to a large family of activating receptor DNAM-1, also called CD226 (6, 7). -

Labinvest201522.Pdf
452A ANNUAL MEETING ABSTRACTS that microRNA-145 (miR-145) expression decreases with the progression of PACa, Design: Resection specimens received in 2012-13 with the diagnosis of DCIS without allowing aberrant expression of MUC13. Restitution of lost levels of miR-145 may IBC were screened. Based on ER, PR, and HER2 status, a total of 18 cases with high prevent PACa progression by down-regulating MUC13 expression. nuclear grade DCIS [6 ER/PR+, HER2–(ER/PR+), 6 ER/PR-, HER2+(HER2), and 6 Design: A pilot study using six-week-old athymic Nu/nu mice was carried out to ER/PR/HER2–(triple negative; TN)] were selected. RNA was extracted from DCIS investigate the effect of miR-145 restitution. Mice were divided into three treatment areas of the formalin fixed paraffin embedded blocks with 6 non-neoplastic breast groups,control (six mice), non-targeting control mimics (NC, six mice) and miR-145 (six tissue controls. Expression array was performed with Affymetrix Human Gene 1.0 ST mice). Tumors in mice were implanted by injecting PACa HPAF-II cells, having high array at Heflin Center for Genomic Science Core Laboratories. The data was analyzed MUC13 expression, into the right flank subcutaneously. After the tumors developed, using the MATLAB software. (avg. volume 80 mm3) miR-145 mice group was injected intratumorally with miR145 Results: A total of 1,682 transcripts were clustered by the specific subtype and mimics 9 times. Tumor growth was followed for 21 days until the tumors reach 700 the difference of their signal intensity. Thirty genes were identified to be under or mm3. -

CD226 T Cells Expressing the Receptors TIGIT and Divergent
Divergent Phenotypes of Human Regulatory T Cells Expressing the Receptors TIGIT and CD226 This information is current as Christopher A. Fuhrman, Wen-I Yeh, Howard R. Seay, Priya of October 1, 2021. Saikumar Lakshmi, Gaurav Chopra, Lin Zhang, Daniel J. Perry, Stephanie A. McClymont, Mahesh Yadav, Maria-Cecilia Lopez, Henry V. Baker, Ying Zhang, Yizheng Li, Maryann Whitley, David von Schack, Mark A. Atkinson, Jeffrey A. Bluestone and Todd M. Brusko J Immunol published online 20 May 2015 Downloaded from http://www.jimmunol.org/content/early/2015/05/20/jimmun ol.1402381 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2015/05/20/jimmunol.140238 Material 1.DCSupplemental Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision by guest on October 1, 2021 • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2015 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published May 20, 2015, doi:10.4049/jimmunol.1402381 The Journal of Immunology Divergent Phenotypes of Human Regulatory T Cells Expressing the Receptors TIGIT and CD226 Christopher A. -

Delineating Spatiotemporal and Hierarchical Development of Human Fetal Innate Lymphoid Cells
www.nature.com/cr Cell Research www.cell-research.com ARTICLE OPEN Delineating spatiotemporal and hierarchical development of human fetal innate lymphoid cells 1,7 2,7 3,7 4,7 2 5 1 1 1 Chen Liu , Yandong Gong , Han Zhang , Hua Yang , Yang Zeng , Zhilei✉ Bian , Qian✉ Xin , Zhijie Bai✉ , Man Zhang , ✉ Jian He 1, Jing Yan1, Jie Zhou2, Zongcheng Li 2, Yanli Ni2, Aiqing Wen3 , Yu Lan 5 , Hongbo Hu 6 and Bing Liu 1,2,5,8 © The Author(s) 2021 Whereas the critical roles of innate lymphoid cells (ILCs) in adult are increasingly appreciated, their developmental hierarchy in early human fetus remains largely elusive. In this study, we sorted human hematopoietic stem/progenitor cells, lymphoid progenitors, putative ILC progenitor/precursors and mature ILCs in the fetal hematopoietic, lymphoid and non-lymphoid tissues, from 8 to 12 post-conception weeks, for single-cell RNA-sequencing, followed by computational analysis and functional validation at bulk and single-cell levels. We delineated the early phase of ILC lineage commitment from hematopoietic stem/progenitor cells, which mainly occurred in fetal liver and intestine. We further unveiled interleukin-3 receptor as a surface marker for the lymphoid progenitors in fetal liver with T, B, ILC and myeloid potentials, while IL-3RA– lymphoid progenitors were predominantly B-lineage committed. Notably, we determined the heterogeneity and tissue distribution of each ILC subpopulation, revealing the proliferating characteristics shared by the precursors of each ILC subtype. Additionally, a novel unconventional ILC2 subpopulation (CRTH2– CCR9+ ILC2) was identified in fetal thymus. Taken together, our study illuminates the precise cellular and molecular features underlying the stepwise formation of human fetal ILC hierarchy with remarkable spatiotemporal heterogeneity. -
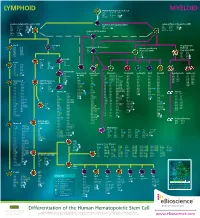
Lymphoid Myeloid
LYMPHOID Human Hematopoietic Stem Cell MYELOID CD34 CD117 (c-kit) CD338 CD38low/neg CD133 linneg CD59 CD135 (Flt3) GATA2 CD90 (Thy1) CD164 TdT Common Lymphoid Progenitor (CLP) Multi-Potent Progenitor (MPP) Common Myeloid Progenitor (CMP) low neg CD33 CD123low CD174 CD7 CD117 (c-kit) C/EBPα CD34 linneg neg CD34 CD131 Ikaros CD10 CD127 GATA2 CD135 (Flt3) TdT CD24neg CD135 (Flt3) GATA3 CD45RA CD164 PU.1 CD34 CD164 PU.1high Common DC Progenitor CD117 (c-kit) CD173 CD38 HLA-DR TdT CD45RA Aiolos CD11c dim CD90low c-mybhigh CD33 Pro-B Pre-NK/T Megakaryocyte- CD10 CD124 CD5 Conventional DC Precursor Erythroid CD19 CD164 CD34 CD11c Granulocyte-Myeloid CD20 CD252 CD14neg Progenitor (GMP) Progenitor (MEP) CD22 CD268 CD33dim CD33 CD131 CD34neg CD275 CD34 CD45RA CD34 CD164 CD72 HLA-DR CD36 FOG-1 CD74 CD123 PU.1 CD110 GATA1 CD123high GATA2 Pre-T Pro-NK CD7 Pre-B CD1a CD28 CD10neg CD2 CD34 neg CD10 CD124 CD34 CD3 CD45 CD117 (c-kit) CD19 CD164 CD5 CD127 (IL-7Rα) CD20 CD252 CD7high NOTCH1 CD22 CD268 CD34neg CD275 CD72 HLA-DR Immature NK CD74 IgM CD34neg Plasmacytoid Conventional Monocyte Neutrophil Eosinophil Mast Basophil Megakaryocyte Erythrocyte CD79a Pax5 CD94neg DC (pDC) DC (cDC) CD4low CD85a (ILT5) CD171 CD10 CD9 CD9 CD9 CD41 CD51 CD35 CD236 CD117 (c-kit) CD9 CD85d (ILT4) CD172a (SIRPα) CD15 CD11b CD11b CD11a CD42a CD61 CD44 CD236R CD4 CD1c CD122 CD11b CD85h (ILT1) CD172b CD16b CD15 CD15 CD11b CD42b CD110 CD123 CD238 CD11c neg CD1d CD161 CD11c CD85j (ILT2) CD180 CD17 CD24 CD24 CD13 CD42c CD112 CD173 CD239 CD45RA CD2 Ets-1 CD13 -

Cell Function the TIGIT/CD226 Axis Regulates Human T
The TIGIT/CD226 Axis Regulates Human T Cell Function Ester Lozano, Margarita Dominguez-Villar, Vijay Kuchroo and David A. Hafler This information is current as of September 26, 2021. J Immunol 2012; 188:3869-3875; Prepublished online 16 March 2012; doi: 10.4049/jimmunol.1103627 http://www.jimmunol.org/content/188/8/3869 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2012/03/16/jimmunol.110362 Material 7.DC1 References This article cites 24 articles, 8 of which you can access for free at: http://www.jimmunol.org/ http://www.jimmunol.org/content/188/8/3869.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 26, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology The TIGIT/CD226 Axis Regulates Human T Cell Function Ester Lozano,*,†,‡ Margarita Dominguez-Villar,*,†,‡ Vijay Kuchroo,‡,1 and David A. Hafler*,†,‡,1 T cell Ig and ITIM domain (TIGIT) is a newly identified receptor expressed on T cells that binds to CD155 on the dendritic cell surface, driving them to a more tolerogenic phenotype. -

Overcoming Challenges for CD3-Bispecific Antibody Therapy In
cancers Review Overcoming Challenges for CD3-Bispecific Antibody Therapy in Solid Tumors Jim Middelburg 1 , Kristel Kemper 2, Patrick Engelberts 2 , Aran F. Labrijn 2 , Janine Schuurman 2 and Thorbald van Hall 1,* 1 Department of Medical Oncology, Oncode Institute, Leiden University Medical Center, 2333 ZA Leiden, The Netherlands; [email protected] 2 Genmab, 3584 CT Utrecht, The Netherlands; [email protected] (K.K.); [email protected] (P.E.); [email protected] (A.F.L.); [email protected] (J.S.) * Correspondence: [email protected]; Tel.: +31-71-5266945 Simple Summary: CD3-bispecific antibody therapy is a form of immunotherapy that enables soldier cells of the immune system to recognize and kill tumor cells. This type of therapy is currently successfully used in the clinic to treat tumors in the blood and is under investigation for tumors in our organs. The treatment of these solid tumors faces more pronounced hurdles, which affect the safety and efficacy of CD3-bispecific antibody therapy. In this review, we provide a brief status update of this field and identify intrinsic hurdles for solid cancers. Furthermore, we describe potential solutions and combinatorial approaches to overcome these challenges in order to generate safer and more effective therapies. Abstract: Immunotherapy of cancer with CD3-bispecific antibodies is an approved therapeutic option for some hematological malignancies and is under clinical investigation for solid cancers. However, the treatment of solid tumors faces more pronounced hurdles, such as increased on-target off-tumor toxicities, sparse T-cell infiltration and impaired T-cell quality due to the presence of an Citation: Middelburg, J.; Kemper, K.; immunosuppressive tumor microenvironment, which affect the safety and limit efficacy of CD3- Engelberts, P.; Labrijn, A.F.; bispecific antibody therapy. -

Mouse CD Marker Chart Bdbiosciences.Com/Cdmarkers
BD Mouse CD Marker Chart bdbiosciences.com/cdmarkers 23-12400-01 CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD Alternative Name Ligands & Associated Molecules T Cell B Cell Dendritic Cell NK Cell Stem Cell/Precursor Macrophage/Monocyte Granulocyte Platelet Erythrocyte Endothelial Cell Epithelial Cell CD1d CD1.1, CD1.2, Ly-38 Lipid, Glycolipid Ag + + + + + + + + CD104 Integrin b4 Laminin, Plectin + DNAX accessory molecule 1 (DNAM-1), Platelet and T cell CD226 activation antigen 1 (PTA-1), T lineage-specific activation antigen 1 CD112, CD155, LFA-1 + + + + + – + – – CD2 LFA-2, Ly-37, Ly37 CD48, CD58, CD59, CD15 + + + + + CD105 Endoglin TGF-b + + antigen (TLiSA1) Mucin 1 (MUC1, MUC-1), DF3 antigen, H23 antigen, PUM, PEM, CD227 CD54, CD169, Selectins; Grb2, β-Catenin, GSK-3β CD3g CD3g, CD3 g chain, T3g TCR complex + CD106 VCAM-1 VLA-4 + + EMA, Tumor-associated mucin, Episialin + + + + + + Melanotransferrin (MT, MTF1), p97 Melanoma antigen CD3d CD3d, CD3 d chain, T3d TCR complex + CD107a LAMP-1 Collagen, Laminin, Fibronectin + + + CD228 Iron, Plasminogen, pro-UPA (p97, MAP97), Mfi2, gp95 + + CD3e CD3e, CD3 e chain, CD3, T3e TCR complex + + CD107b LAMP-2, LGP-96, LAMP-B + + Lymphocyte antigen 9 (Ly9), -

New Weapons for Natural Killer Cell Arsenal Against Hematological Malignancies
cells Review Checkpoint Inhibitors and Engineered Cells: New Weapons for Natural Killer Cell Arsenal Against Hematological Malignancies Massimo Giuliani 1,* and Alessandro Poggi 2 1 Department of Oncology, Luxembourg Institute of Health, Luxembourg City L-1526, Luxembourg 2 Molecular Oncology and Angiogenesis Unit, IRCCS AOU San Martino IST, 16132 Genoa, Italy; [email protected] * Correspondence: [email protected] Received: 1 June 2020; Accepted: 25 June 2020; Published: 29 June 2020 Abstract: Natural killer (NK) cells represent one of the first lines of defense against malignant cells. NK cell activation and recognition are regulated by a balance between activating and inhibitory receptors, whose specific ligands can be upregulated on tumor cells surface and tumor microenvironment (TME). Hematological malignancies set up an extensive network of suppressive factors with the purpose to induce NK cell dysfunction and impaired immune-surveillance ability. Over the years, several strategies have been developed to enhance NK cells-mediated anti-tumor killing, while other approaches have arisen to restore the NK cell recognition impaired by tumor cells and other cellular components of the TME. In this review, we summarize and discuss the strategies applied in hematological malignancies to block the immune check-points and trigger NK cells anti-tumor effects through engineered chimeric antigen receptors. Keywords: NK cells; hematological malignancies; check-point inhibitors; CAR NK cells; antibodies; immunotherapy 1. Tumor-Mediated NK Cell Exhaustion in Hematological Malignancies NK cells represent one of the first lines of defense against malignant cells. Their immune- surveillance ability is mediated by the expression of activating receptors such as the natural killer group (NKG)2D, DNAX accessory molecule (DNAM)-1, and the natural cytotoxic receptors (NCRs) such as natural killer protein (NKp)30, NKp44, and NKp46 [1–5]. -

Immune Checkpoint Receptors
T Cell NK Cell Tumor Cell Killing of cancer cells Recognition of cancer cells by T cells NK Cell Tumor Cell T Cell TRAIL TRAILR1/2 ICOSL ICOS NKp30a,b B7-H6/BAT3/BAG6 HHLA2 TMIGD2 NKG2C HLA-E HVEM LIGHT NKG2D MICA family MHC class I/II KIRS NKG2D RAET1/ULBP family CD137L CD137 KIRS MHC class I/II OX40L OX40 CD27 CD70 CD70 CD27 Activation CD226 CD122 GITRL GITR Immune Checkpoint Receptors CD226 CD155 CD155 CD226 Activation CD244 CD48 CD48 CD244 CD86 CD28 CD80 CD28 CD244 CD48 CD96 CD122 CD86 CTLA-4 TIGIT CD122 CD80 CTLA-4 CD96 CD155 CD80 PDL1 TIGIT CD155 PDL1 CD80 NKG2A HLA-E PDL1 PD1 Inhibition PDL2 PD1 HVEM CD272 HVEM CD160 MHC class I/II KIR L Inhibition MHC class I/II CD223 CD155 CD96 CD112 TIGIT CD112 CD112R CD200 CD200R Gal-9 TIM-3 Abnova Abnova Corporation 9F, No. 108, Jhouzih St., Neihu, Taipei 114, Taiwan Tel: + 886 2 8751 1888 Fax: + 886 2 6602 1218 www.abnova.com Catalog No. BAT3 Related Products Applications Reactivity H00007917-AP21 BAT3 (Human) Matched Antibody Pair EPair-Re Human H00007917-P01 BAT3 (Human) Recombinant Protein (P01) AP,Array,ELISA,WB-Re H00007917-T01 BAT3 293T Cell Transient Overexpression Lysate(Denatured) WB H00007917-B01 BAT3 MaxPab mouse polyclonal antibody (B01) WB-Ce,IF,WB-Tr Human H00007917-M03A BAT3 monoclonal antibody (M03A), clone 2B9 ELISA Human H00007917-B01P BAT3 purified MaxPab mouse polyclonal antibody (B01P) WB-Ce,IF,WB-Tr Human Catalog No. BTLA Related Products Applications Reactivity H00151888-Q01 BTLA (Human) Recombinant Protein (Q01) AP,Array,ELISA,WB-Re H00151888-M02 BTLA monoclonal antibody (M02), clone 1B7 ELISA,WB-Re Human H00151888-R01 BTLA Pre-design Chimera RNAi RNAi Human Catalog No.