RNA Structure and Folding Overview
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Comparison of the Effects on Mrna and Mirna Stability Arian Aryani and Bernd Denecke*
Aryani and Denecke BMC Research Notes (2015) 8:164 DOI 10.1186/s13104-015-1114-z RESEARCH ARTICLE Open Access In vitro application of ribonucleases: comparison of the effects on mRNA and miRNA stability Arian Aryani and Bernd Denecke* Abstract Background: MicroRNA has become important in a wide range of research interests. Due to the increasing number of known microRNAs, these molecules are likely to be increasingly seen as a new class of biomarkers. This is driven by the fact that microRNAs are relatively stable when circulating in the plasma. Despite extensive analysis of mechanisms involved in microRNA processing, relatively little is known about the in vitro decay of microRNAs under defined conditions or about the relative stabilities of mRNAs and microRNAs. Methods: In this in vitro study, equal amounts of total RNA of identical RNA pools were treated with different ribonucleases under defined conditions. Degradation of total RNA was assessed using microfluidic analysis mainly based on ribosomal RNA. To evaluate the influence of the specific RNases on the different classes of RNA (ribosomal RNA, mRNA, miRNA) ribosomal RNA as well as a pattern of specific mRNAs and miRNAs was quantified using RT-qPCR assays. By comparison to the untreated control sample the ribonuclease-specific degradation grade depending on the RNA class was determined. Results: In the present in vitro study we have investigated the stabilities of mRNA and microRNA with respect to the influence of ribonucleases used in laboratory practice. Total RNA was treated with specific ribonucleases and the decay of different kinds of RNA was analysed by RT-qPCR and miniaturized gel electrophoresis. -

Exploring the Structure of Long Non-Coding Rnas, J
IMF YJMBI-63988; No. of pages: 15; 4C: 3, 4, 7, 8, 10 1 2 Rise of the RNA Machines: Exploring the Structure of 3 Long Non-Coding RNAs 4 Irina V. Novikova, Scott P. Hennelly, Chang-Shung Tung and Karissa Y. Sanbonmatsu Q15 6 Los Alamos National Laboratory, Los Alamos, NM 87545, USA 7 Correspondence to Karissa Y. Sanbonmatsu: [email protected] 8 http://dx.doi.org/10.1016/j.jmb.2013.02.030 9 Edited by A. Pyle 1011 12 Abstract 13 Novel, profound and unexpected roles of long non-coding RNAs (lncRNAs) are emerging in critical aspects of 14 gene regulation. Thousands of lncRNAs have been recently discovered in a wide range of mammalian 15 systems, related to development, epigenetics, cancer, brain function and hereditary disease. The structural 16 biology of these lncRNAs presents a brave new RNA world, which may contain a diverse zoo of new 17 architectures and mechanisms. While structural studies of lncRNAs are in their infancy, we describe existing 18 structural data for lncRNAs, as well as crystallographic studies of other RNA machines and their implications 19 for lncRNAs. We also discuss the importance of dynamics in RNA machine mechanism. Determining 20 commonalities between lncRNA systems will help elucidate the evolution and mechanistic role of lncRNAs in 21 disease, creating a structural framework necessary to pursue lncRNA-based therapeutics. 22 © 2013 Published by Elsevier Ltd. 24 23 25 Introduction rather than the exception in the case of eukaryotic 50 organisms. 51 26 RNA is primarily known as an intermediary in gene LncRNAs are defined by the following: (i) lack of 52 11 27 expression between DNA and proteins. -

The Architecture of the Protein Domain Universe
The architecture of the protein domain universe Nikolay V. Dokholyan Department of Biochemistry and Biophysics, The University of North Carolina at Chapel Hill, School of Medicine, Chapel Hill, NC 27599 ABSTRACT Understanding the design of the universe of protein structures may provide insights into protein evolution. We study the architecture of the protein domain universe, which has been found to poses peculiar scale-free properties (Dokholyan et al., Proc. Natl. Acad. Sci. USA 99: 14132-14136 (2002)). We examine the origin of these scale-free properties of the graph of protein domain structures (PDUG) and determine that that the PDUG is not modular, i.e. it does not consist of modules with uniform properties. Instead, we find the PDUG to be self-similar at all scales. We further characterize the PDUG architecture by studying the properties of the hub nodes that are responsible for the scale-free connectivity of the PDUG. We introduce a measure of the betweenness centrality of protein domains in the PDUG and find a power-law distribution of the betweenness centrality values. The scale-free distribution of hubs in the protein universe suggests that a set of specific statistical mechanics models, such as the self-organized criticality model, can potentially identify the principal driving forces of molecular evolution. We also find a gatekeeper protein domain, removal of which partitions the largest cluster into two large sub- clusters. We suggest that the loss of such gatekeeper protein domains in the course of evolution is responsible for the creation of new fold families. INTRODUCTION The principles of molecular evolution remain elusive despite fundamental breakthroughs on the theoretical front 1-5 and a growing amount of genomic and proteomic data, over 23,000 solved protein structures 6 and protein functional annotations 7-9. -

RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases
G C A T T A C G G C A T genes Review RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases Amber Willbanks, Shaun Wood and Jason X. Cheng * Department of Pathology, Hematopathology Section, University of Chicago, Chicago, IL 60637, USA; [email protected] (A.W.); [email protected] (S.W.) * Correspondence: [email protected] Abstract: Chromatin structure plays an essential role in eukaryotic gene expression and cell identity. Traditionally, DNA and histone modifications have been the focus of chromatin regulation; however, recent molecular and imaging studies have revealed an intimate connection between RNA epigenetics and chromatin structure. Accumulating evidence suggests that RNA serves as the interplay between chromatin and the transcription and splicing machineries within the cell. Additionally, epigenetic modifications of nascent RNAs fine-tune these interactions to regulate gene expression at the co- and post-transcriptional levels in normal cell development and human diseases. This review will provide an overview of recent advances in the emerging field of RNA epigenetics, specifically the role of RNA modifications and RNA modifying proteins in chromatin remodeling, transcription activation and RNA processing, as well as translational implications in human diseases. Keywords: 5’ cap (5’ cap); 7-methylguanosine (m7G); R-loops; N6-methyladenosine (m6A); RNA editing; A-to-I; C-to-U; 2’-O-methylation (Nm); 5-methylcytosine (m5C); NOL1/NOP2/sun domain Citation: Willbanks, A.; Wood, S.; (NSUN); MYC Cheng, J.X. RNA Epigenetics: Fine-Tuning Chromatin Plasticity and Transcriptional Regulation, and the Implications in Human Diseases. Genes 2021, 12, 627. -

SARS-Cov-2 RNA, Qualitative Real-Time RT-PCR (Test Code 39433)
SARS-CoV-2 RNA, Qualitative Real-Time RT-PCR (Test Code 39433) Package Insert For Emergency Use Only For In-vitro Diagnostic Use - Rx Only Intended Use The Quest Diagnostics SARS-CoV-2 RNA, Qualitative Real-Time RT-PCR (“Quest SARS-CoV-2 rRT-PCR”) is a real-time RT-PCR test intended for the qualitative detection of nucleic acid from the SARS-CoV-2 in upper and lower respiratory specimens (such as nasopharyngeal or oropharyngeal swabs, sputum, tracheal aspirates, and bronchoalveolar lavage) collected from individuals suspected of COVID-19 by their healthcare provider. This test is also for use with nasal swab specimens that are self-collected at home or in a healthcare setting by individuals using an authorized home-collection kit when determined to be appropriate by a healthcare provider. This test is for the qualitative detection of nucleic acid from the SARS-CoV-2 in pooled samples containing up to four of the individual upper respiratory swab specimens (nasopharyngeal, mid-turbinate, anterior nares or oropharyngeal swabs) that were collected in individual vials containing transport media from individuals suspected of COVID-19 by their healthcare provider. Negative results from pooled testing should not be treated as definitive. If patient’s clinical signs and symptoms are inconsistent with a negative result or results are necessary for patient management, then the patient should be considered for individual testing. Specimens included in pools with a positive, inconclusive, or invalid result must be tested individually prior to reporting a result. Specimens with low viral loads may not be detected in sample pools due to the decreased sensitivity of pooled testing. -

Split Deoxyribozyme Probe for Efficient Detection of Highly Structured RNA Targets
University of Central Florida STARS Honors Undergraduate Theses UCF Theses and Dissertations 2018 Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets Sheila Raquel Solarez University of Central Florida Part of the Biochemistry Commons, and the Biology Commons Find similar works at: https://stars.library.ucf.edu/honorstheses University of Central Florida Libraries http://library.ucf.edu This Open Access is brought to you for free and open access by the UCF Theses and Dissertations at STARS. It has been accepted for inclusion in Honors Undergraduate Theses by an authorized administrator of STARS. For more information, please contact [email protected]. Recommended Citation Solarez, Sheila Raquel, "Split Deoxyribozyme Probe For Efficient Detection of Highly Structured RNA Targets" (2018). Honors Undergraduate Theses. 311. https://stars.library.ucf.edu/honorstheses/311 SPLIT DEOXYRIBOZYME PROBE FOR EFFICIENT DETECTION OF HIGHLY STRUCTURED RNA TARGETS By SHEILA SOLAREZ A thesis submitted in partial fulfillment of the requirements for the Honors in the Major Program in Biological Sciences in the College of Sciences and the Burnett Honors College at the University of Central Florida Orlando, Florida Spring Term, 2018 Thesis Chair: Yulia Gerasimova, PhD ABSTRACT Transfer RNAs (tRNAs) are known for their role as adaptors during translation of the genetic information and as regulators for gene expression; uncharged tRNAs regulate global gene expression in response to changes in amino acid pools in the cell. Aminoacylated tRNAs play a role in non-ribosomal peptide bond formation, post-translational protein labeling, modification of phospholipids in the cell membrane, and antibiotic biosynthesis. [1] tRNAs have a highly stable structure that can present a challenge for their detection using conventional techniques. -

The Difficult Case of an RNA-Only Origin of Life
Emerging Topics in Life Sciences (2019) 3 469–475 https://doi.org/10.1042/ETLS20190024 Perspective The difficult case of an RNA-only origin of life Kristian Le Vay and Hannes Mutschler Max-Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany Downloaded from https://portlandpress.com/emergtoplifesci/article-pdf/3/5/469/859756/etls-2019-0024c.pdf by Max-Planck-Institut fur Biochemie user on 28 November 2019 Correspondence: Hannes Mutschler ([email protected]) The RNA world hypothesis is probably the most extensively studied model for the emergence of life on Earth. Despite a large body of evidence supporting the idea that RNA is capable of kick-starting autocatalytic self-replication and thus initiating the emergence of life, seemingly insurmountable weaknesses in the theory have also been highlighted. These problems could be overcome by novel experimental approaches, including out-of- equilibrium environments, and the exploration of an early co-evolution of RNA and other key biomolecules such as peptides and DNA, which might be necessary to mitigate the shortcomings of RNA-only systems. The conjecture that life on Earth evolved from an ‘RNA World’ remains one of the most popular hypotheses for abiogenesis, even 60 years after Alex Rich first put the idea forward [1]. For some, evi- dence based upon ubiquitous molecular fossils and the elegance of the idea that RNA once had a dual role as information carrier and prebiotic catalyst provide overwhelming support for the theory. Nevertheless, doubts remain surrounding the chemical evolution of an RNA world, whose classical scenario is based on a temporal sequence of nucleotide formation, enzyme-free polymerisation/replica- tion, recombination, encapsulation in lipid vesicles (or other compartments), evolution of ribozymes and finally the innovation of the genetic code and its translation (Figure 1)[2,3]. -

Mrna Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection
International Journal of Molecular Sciences Review mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection 1, 1, 2 1,3, Shuqin Xu y, Kunpeng Yang y, Rose Li and Lu Zhang * 1 State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Science, Fudan University, Shanghai 200438, China; [email protected] (S.X.); [email protected] (K.Y.) 2 M.B.B.S., School of Basic Medical Sciences, Peking University Health Science Center, Beijing 100191, China; [email protected] 3 Shanghai Engineering Research Center of Industrial Microorganisms, Shanghai 200438, China * Correspondence: [email protected]; Tel.: +86-13524278762 These authors contributed equally to this work. y Received: 30 July 2020; Accepted: 30 August 2020; Published: 9 September 2020 Abstract: Messenger ribonucleic acid (mRNA)-based drugs, notably mRNA vaccines, have been widely proven as a promising treatment strategy in immune therapeutics. The extraordinary advantages associated with mRNA vaccines, including their high efficacy, a relatively low severity of side effects, and low attainment costs, have enabled them to become prevalent in pre-clinical and clinical trials against various infectious diseases and cancers. Recent technological advancements have alleviated some issues that hinder mRNA vaccine development, such as low efficiency that exist in both gene translation and in vivo deliveries. mRNA immunogenicity can also be greatly adjusted as a result of upgraded technologies. In this review, we have summarized details regarding the optimization of mRNA vaccines, and the underlying biological mechanisms of this form of vaccines. Applications of mRNA vaccines in some infectious diseases and cancers are introduced. It also includes our prospections for mRNA vaccine applications in diseases caused by bacterial pathogens, such as tuberculosis. -

Expanding the Genetic Code Lei Wang and Peter G
Reviews P. G. Schultz and L. Wang Protein Science Expanding the Genetic Code Lei Wang and Peter G. Schultz* Keywords: amino acids · genetic code · protein chemistry Angewandte Chemie 34 2005 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim DOI: 10.1002/anie.200460627 Angew. Chem. Int. Ed. 2005, 44,34–66 Angewandte Protein Science Chemie Although chemists can synthesize virtually any small organic molecule, our From the Contents ability to rationally manipulate the structures of proteins is quite limited, despite their involvement in virtually every life process. For most proteins, 1. Introduction 35 modifications are largely restricted to substitutions among the common 20 2. Chemical Approaches 35 amino acids. Herein we describe recent advances that make it possible to add new building blocks to the genetic codes of both prokaryotic and 3. In Vitro Biosynthetic eukaryotic organisms. Over 30 novel amino acids have been genetically Approaches to Protein encoded in response to unique triplet and quadruplet codons including Mutagenesis 39 fluorescent, photoreactive, and redox-active amino acids, glycosylated 4. In Vivo Protein amino acids, and amino acids with keto, azido, acetylenic, and heavy-atom- Mutagenesis 43 containing side chains. By removing the limitations imposed by the existing 20 amino acid code, it should be possible to generate proteins and perhaps 5. An Expanded Code 46 entire organisms with new or enhanced properties. 6. Outlook 61 1. Introduction The genetic codes of all known organisms specify the same functional roles to amino acid residues in proteins. Selectivity 20 amino acid building blocks. These building blocks contain a depends on the number and reactivity (dependent on both limited number of functional groups including carboxylic steric and electronic factors) of a particular amino acid side acids and amides, a thiol and thiol ether, alcohols, basic chain. -

RNA Quality Control in Mirna Expression Analysis
RNA Quality Control in miRNA Expression Analysis Application Note Genomics Authors Abstract Christiane Becker, Michael W. Pfaffl It is generally known that total RNA quality has a distinct influence on the validity and TU München, ZIEL reliability of quantitative PCR results. In addition, the recently published MIQE Physiology Weihenstephan guidelines focus on the pre-PCR steps and state the importance of RNA quality Weihenstephaner Berg 3 assessment. 85384 Freising, Germany Various studies showed the impairing effect of ongoing RNA degradation on mRNA Martina Reiter, Michael W. Pfaffl expression results. Therefore, the verification of RNA integrity prior to downstream BioEPS GmbH, Lise-Meitner-Str. 30 applications like RT-qPCR and mircroarrays is indispensable. A fast and reliable 85354 Freising, Germany assessment of RNA integrity can be done with the Eukaryote Total RNA Nano Assay of the Agilent 2100 Bioanalyzer System. The importance of RNA quality should also be considered in new applications such as the investigation of miRNA expression profiles. With the Agilent Small RNA Assay, Agilent is offering one of the few possibilities for selectively estimating miRNA before expression analysis. However, by now little is known about factors affecting miRNA analysis. Herein, the important impact of total RNA quality on quantification of mRNA and miRNA should be considered. Introduction After extraction, RNAs are very unsta- ble and sensitive to degradation due to 1A the ubiquitous occurrence of nucleases. It is well known that expression profil- ing using microarrays or qPCR is influ- enced by RNA quality1, 2. Therefore, this hazard has to be considered during all preprocessing steps and should be min- imized with cautious handling. -

Cmse 520 Biomolecular Structure, Function And
CMSE 520 BIOMOLECULAR STRUCTURE, FUNCTION AND DYNAMICS (Computational Structural Biology) OUTLINE Review: Molecular biology Proteins: structure, conformation and function(5 lectures) Generalized coordinates, Phi, psi angles, DNA/RNA: structure and function (3 lectures) Structural and functional databases (PDB, SCOP, CATH, Functional domain database, gene ontology) Use scripting languages (e.g. python) to cross refernce between these databases: starting from sequence to find the function Relationship between sequence, structure and function Molecular Modeling, homology modeling Conservation, CONSURF Relationship between function and dynamics Confromational changes in proteins (structural changes due to ligation, hinge motions, allosteric changes in proteins and consecutive function change) Molecular Dynamics Monte Carlo Protein-protein interaction: recognition, structural matching, docking PPI databases: DIP, BIND, MINT, etc... References: CURRENT PROTOCOLS IN BIOINFORMATICS (e-book) (http://www.mrw.interscience.wiley.com/cp/cpbi/articles/bi0101/frame.html) Andreas D. Baxevanis, Daniel B. Davison, Roderic D.M. Page, Gregory A. Petsko, Lincoln D. Stein, and Gary D. Stormo (eds.) 2003 John Wiley & Sons, Inc. INTRODUCTION TO PROTEIN STRUCTURE Branden C & Tooze, 2nd ed. 1999, Garland Publishing COMPUTER SIMULATION OF BIOMOLECULAR SYSTEMS Van Gusteren, Weiner, Wilkinson Internet sources Ref: Department of Energy Rapid growth in experimental technologies Human Genome Projects Two major goals 1. DNA mapping 2. DNA sequencing Rapid growth in experimental technologies z Microrarray technologies – serial gene expression patterns and mutations z Time-resolved optical, rapid mixing techniques - folding & function mechanisms (Æ ns) z Techniques for probing single molecule mechanics (AFM, STM) (Æ pN) Æ more accurate models/data for computer-aided studies Weiss, S. (1999). Fluorescence spectroscopy of single molecules. -
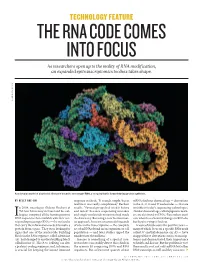
THE RNA CODE COMES INTO FOCUS As Researchers Open up to the Reality of RNA Modification, an Expanded Epitranscriptomics Toolbox Takes Shape
TECHNOLOGY FEATURE THE RNA CODE COMES INTO FOCUS As researchers open up to the reality of RNA modification, an expanded epitranscriptomics toolbox takes shape. LAGUNA DESIGN/SPL LAGUNA A molecular model of a bacterial ribosome bound to messenger RNA, a complex that is formed during protein synthesis. BY KELLY RAE CHI response in check. “It sounds simple, but in mRNAs harbour chemical tags — decorations real life it was really complicated,” Rechavi to the A, C, G and U nucleotides — that are n 2004, oncologist Gideon Rechavi at recalls. “Several groups had tried it before invisible to today’s sequencing technologies. Tel Aviv University in Israel and his col- and failed” because sequencing mistakes (Similar chemical tags, called epigenetic mark- leagues compared all the human genomic and single-nucleotide mutations had made ers, are also found on DNA.) Researchers aren’t IDNA sequences then available with their cor- the data noisy. But using a new bioinformat- sure what these chemical changes in RNA do, responding messenger RNAs — the molecules ics approach, his team uncovered thousands but they’re trying to find out. that carry the information needed to make a of sites in the transcriptome — the complete A wave of studies over the past five years — protein from a gene. They were looking for set of mRNAs found in an organism or cell many of which focus on a specific RNA mark signs that one of the nucleotide building population — and later studies upped the called N6-methyladenosine (m6A) — have blocks in the RNA sequence, called adenosine number into the millions1.