Isolation and Characterization of Microsatellite Marker Loci in The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Classification of Mammals 61
© Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FORCHAPTER SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION Classification © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC 4 NOT FORof SALE MammalsOR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC © Jones & Bartlett Learning, LLC NOT FOR SALE OR DISTRIBUTION NOT FOR SALE OR DISTRIBUTION © Jones & Bartlett Learning, LLC. NOT FOR SALE OR DISTRIBUTION. 2ND PAGES 9781284032093_CH04_0060.indd 60 8/28/13 12:08 PM CHAPTER 4: Classification of Mammals 61 © Jones Despite& Bartlett their Learning,remarkable success, LLC mammals are much less© Jones stress & onBartlett the taxonomic Learning, aspect LLCof mammalogy, but rather as diverse than are most invertebrate groups. This is probably an attempt to provide students with sufficient information NOT FOR SALE OR DISTRIBUTION NOT FORattributable SALE OR to theirDISTRIBUTION far greater individual size, to the high on the various kinds of mammals to make the subsequent energy requirements of endothermy, and thus to the inabil- discussions of mammalian biology meaningful. -

Diet and the Evolution of Digestion and Renal Function in Phyllostomid Bats
Zoology 104 (2001): 59–73 © by Urban & Fischer Verlag http://www.urbanfischer.de/journals/zoology Diet and the evolution of digestion and renal function in phyllostomid bats Jorge E. Schondube1,*, L. Gerardo Herrera-M.2 and Carlos Martínez del Rio1 1Department of Ecology and Evolutionary Biology, University of Arizona, Tucson 2Instituto de Biología, Departamento de Zoología, Universidad Nacional Autónoma de México, México Received: April 2, 2001 · Accepted: April 12, 2001 Abstract Bat species in the monophyletic family Phyllostomidae feed on blood, insects, small vertebrates, nectar, fruit and complex omnivorous mixtures. We used nitrogen stable isotope ratios to characterize bat diets and adopted a phylogenetically informed approach to investi- gate the physiological changes that accompany evolutionary diet changes in phyllostomids. We found that nitrogen stable isotopes sep- arated plant-eating from animal-eating species. The blood of the latter was enriched in 15N. A recent phylogenetic hypothesis suggests that with the possible exception of carnivory, which may have evolved twice, all diets evolved only once from insectivory. The shift from insectivory to nectarivory and frugivory was accompanied by increased intestinal sucrase and maltase activity, decreased trehalase activity, and reduced relative medullary thickness of kidneys. The shift from insectivory to sanguinivory and carnivory resulted in re- duced trehalase activity. Vampire bats are the only known vertebrates that do not exhibit intestinal maltase activity. We argue that these physiological changes are adaptive responses to evolutionary diet shifts. Key words: Bats, comparative method, diet, digestive and renal function, stable isotopes. Introduction The family Phyllostomidae is a speciose (49 genera and Characterizing animal diets can be difficult. -

BATS of the Golfo Dulce Region, Costa Rica
MURCIÉLAGOS de la región del Golfo Dulce, Puntarenas, Costa Rica BATS of the Golfo Dulce Region, Costa Rica 1 Elène Haave-Audet1,2, Gloriana Chaverri3,4, Doris Audet2, Manuel Sánchez1, Andrew Whitworth1 1Osa Conservation, 2University of Alberta, 3Universidad de Costa Rica, 4Smithsonian Tropical Research Institute Photos: Doris Audet (DA), Joxerra Aihartza (JA), Gloriana Chaverri (GC), Sébastien Puechmaille (SP), Manuel Sánchez (MS). Map: Hellen Solís, Universidad de Costa Rica © Elène Haave-Audet [[email protected]] and other authors. Thanks to: Osa Conservation and the Bobolink Foundation. [fieldguides.fieldmuseum.org] [1209] version 1 11/2019 The Golfo Dulce region is comprised of old and secondary growth seasonally wet tropical forest. This guide includes representative species from all families encountered in the lowlands (< 400 masl), where ca. 75 species possibly occur. Species checklist for the region was compiled based on bat captures by the authors and from: Lista y distribución de murciélagos de Costa Rica. Rodríguez & Wilson (1999); The mammals of Central America and Southeast Mexico. Reid (2012). Taxonomy according to Simmons (2005). La región del Golfo Dulce está compuesta de bosque estacionalmente húmedo primario y secundario. Esta guía incluye especies representativas de las familias presentes en las tierras bajas de la región (< de 400 m.s.n.m), donde se puede encontrar c. 75 especies. La lista de especies fue preparada con base en capturas de los autores y desde: Lista y distribución de murciélagos de Costa Rica. Rodríguez -
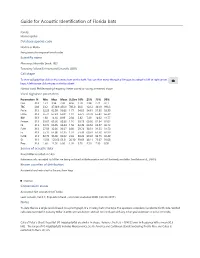
Mormoops Blainvillei Leach, 1821 Taxonomy Follows Simmons and Cirranello (2021) Call Shape
Guide for Acoustic Identification of Florida bats Family: Mormoopidae Database species code Morbla or Moba See glossary for explanation of codes Scientific name Mormoops blainvillei Leach, 1821 Taxonomy follows Simmons and Cirranello (2021) Call shape To view call graphics click on the camera icon on the right. You can then move through all images by using the left or right arrow keys. A left mouse click returns to the fact sheet. Narrow band, FM decreasing frequency. Never paired or having a reverse J shape Vocal signature parameters Parameters N Min Max Mean St.Dev 10% 25% 75% 90% Dur 313 1.51 3.94 2.36 0.56 1.70 1.94 2.71 3.17 TBC 238 33.2 4730.9 456.9 785.0 60.0 122.2 333.9 993.3 Fmin 313 52.29 62.99 56.03 1.77 54.05 54.61 57.35 58.39 Fmax 313 56.74 67.23 65.01 1.47 63.75 64.78 65.84 66.39 BW 313 1.38 14.10 8.99 2.30 5.82 7.49 10.62 11.77 Fmean 313 55.67 63.66 60.86 1.16 59.73 60.36 61.54 62.02 Fk 313 56.74 66.95 64.84 1.50 63.49 64.52 65.57 66.12 FcH1 313 27.59 32.00 30.67 0.80 29.74 30.31 31.25 31.50 Fc 313 55.17 64.00 61.35 1.59 59.48 60.61 62.50 62.99 FcH3 313 82.76 96.00 92.02 2.39 89.22 90.92 93.75 94.49 Sc 313 15.56 125.66 63.51 20.30 39.69 48.11 75.67 91.08 Pmc 313 1.60 14.50 6.00 1.95 3.70 4.50 7.30 8.50 Source of acoustic data Bruce Miller recorded in Cuba Reference calls recorded by Miller are being archived at BioAcoustica and will be freely available. -

Research Article Isolation and Characterization of Microsatellite
Research Article Isolation and characterization of microsatellite marker loci in the Wagner's mustached bat Pteronotus psilotis (Chiroptera: Mormoopidae) and cross-amplification in other related species A. MÉNDEZ-RODRÍGUEZ1, R. LÓPEZ-WILCHIS1*, A. SERRATO DÍAZ2, J. JUSTE3,5, M. A. DEL RÍO- PORTILLA4, L. M. GUEVARA-CHUMACERO1* 1Departamento de Biología, Universidad Autónoma Metropolitana Iztapalapa, Avenida San Rafael Atlixco No. 186, Col. Vicentina. Del. Iztapalapa C.P. 09340, México, Cuidad de México, México 2Departamento de Hidrobiología, Universidad Autónoma Metropolitana Iztapalapa, Avenida San Rafael Atlixco No. 186, Col. Vicentina. Del. Iztapalapa C.P. 09340, México, Cuidad de México, México 3Estación Biológica de Doñana, C.S.I.C., Avda. Américo Vespucio 26, 41092 Sevilla, España 4Departamento de Acuicultura, Centro de Investigación Científica y de Educación Superior de Ensenada, B.C., Carretera Ensenada-Tijuana #3918, Zona Playitas, Ensenada, B. C., C.P. 22860, México 5CIBER de Epidemiología y Salud Pública, CIBERESP, España *For correspondence. Departamento de Biología, Universidad Autónoma Metropolitana Iztapalapa, Avenida San Rafael Atlixco No. 186, Col. Vicentina. Del. Iztapalapa C.P. 09340, México, Distrito Federal, México E-mail address: [email protected] (L. M. Guevara-Chumacero) [email protected] (R. López-Wilchis) Abstract Pteronotus psilotis, a mormoopid bat, is an insectivorous, gregarious and strict cave-dwelling species that is found from sea level to an elevation of about 1,000 m asl. The species is present in diverse habitats ranging from rain forest to dry deciduous forest. Nine microsatellite loci were developed for Wagner's mustached bat P. psilotis using a next-generation sequencing approach, and their utility for population genetics studies was 1 assessed. -

Genetic Introgression and Morphological Variation in Naked-Back Bats (Chiroptera: Mormoopidae: Pteronotus Species) Along Their Contact Zone in Central America
diversity Article Genetic Introgression and Morphological Variation in Naked-Back Bats (Chiroptera: Mormoopidae: Pteronotus Species) along Their Contact Zone in Central America Aline Méndez-Rodríguez 1, Javier Juste 2,3,* , Alejandro Centeno-Cuadros 4 , Flor Rodríguez-Gómez 5, Alejandra Serrato-Díaz 6, Juan Luis García-Mudarra 2, Luis Manuel Guevara-Chumacero 1 and Ricardo López-Wilchis 1,* 1 Departamento de Biología, Universidad Autónoma Metropolitana-Iztapalapa, Av. San Rafael Atlixco 186, Col. Vicentina, Ciudad de Mexico 09340, Mexico; [email protected] (A.M.-R.); [email protected] (L.M.G.-C.) 2 Estación Biológica de Doñana, C.S.I.C., Avda. Américo Vespucio 26, 41092 Sevilla, Spain; [email protected] 3 CIBER de Epidemiología y Salud Pública, CIBERESP, 28220 Madrid, Spain 4 INMAR, Facultad de Ciencias del Mar y Ambientales, Biomedicina, Biotecnologia y Salud Pública, Universidad de Cádiz, 11510 Cádiz, Spain; [email protected] 5 Centro Universitario de Ciencias Exactas e Ingenierías, Departamento de Ciencias Computacionales, Universidad de Guadalajara, Blvd. Gral., Marcelino García Barragán 1421, Olímpica, Citation: Méndez-Rodríguez, A.; Guadalajara 44430, Mexico; fiores.fl[email protected] 6 Juste, J.; Centeno-Cuadros, A.; Departamento de Hidrobiología, Universidad Autónoma Metropolitana-Iztapalapa, Av. San Rafael Atlixco Rodríguez-Gómez, F.; Serrato-Díaz, 186, Col. Vicentina, Ciudad de Mexico 09340, Mexico; [email protected] * Correspondence: [email protected] (J.J.); [email protected] (R.L.-W.) A.; García-Mudarra, J.L.; Guevara-Chumacero, L.M.; López-Wilchis, R. Genetic Abstract: Two sibling bare-backed bat species (Pteronotus fulvus and P. gymnonotus) have been tra- Introgression and Morphological ditionally differentiated by their size. -

Boreal Mammalogy 2019 Syllabus
BOREAL MAMMALOGY ACM Wilderness Field Station Program Long Course Description The behavior, ecology and morphology of animals have been shaped by evolution, within the constraints of anatomy and physiology. Thus animals are adapted in many ways to the environments around them. In this course, we shall investigate the adaptations of mammals for life in the Quetico/Superior region. Most mammals are secretive and more difficult to observe than some other animals, such as birds and insects. Mammals, however, often leave evidence of their activities: tracks and scats (feces) are often easy to find, scrapings associated with scent marks are often left on the ground or trees, well used trails can be found in the woods, and plants eaten by mammals have missing leaves, branches or bark. Many small mammals are easy to live-trap for studies of behavior, population dynamics and reproduction. And some mammals, such as squirrels and beavers, can be observed directly, if one has patience. Within the course of a month there is much that can be learned about the mammals of the Quetico/Superior. The main topics for investigation in this course will be mammal life histories, foods, habitat choices and effects on habitat, populations, and natural history. We shall set up live-trapping grids to study small mammals, take hikes to find tracks, scats, animal trails, take day-trips by canoe and longer trips to places we can not reach on foot and that will give us opportunities not available near the field station to see mammals and their sign. We may also do some small experiments around beaver ponds. -

Species Diversity and Phylogeny of Mustached Bats (Mormoopidae
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/305362807 Integrating Multiples Evidences in Taxonomy: Species Diversity and Phylogeny of Mustached Bats (Mormoopidae... Article in Molecular Phylogenetics and Evolution · July 2016 DOI: 10.1016/j.ympev.2016.07.011 CITATIONS READS 0 228 2 authors, including: Ana Pavan University of São Paulo 8 PUBLICATIONS 75 CITATIONS SEE PROFILE Some of the authors of this publication are also working on these related projects: Systematics of Family Mormoopidae (Chiroptera) View project All content following this page was uploaded by Ana Pavan on 27 July 2016. The user has requested enhancement of the downloaded file. All in-text references underlined in blue are added to the original document and are linked to publications on ResearchGate, letting you access and read them immediately. Molecular Phylogenetics and Evolution xxx (2016) xxx–xxx Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Integrating multiple evidences in taxonomy: species diversity and phylogeny of mustached bats (Mormoopidae: Pteronotus) ⇑ Ana Carolina Pavan ,1, Gabriel Marroig Departamento de Genética e Biologia Evolutiva, IB/Universidade de São Paulo, 05508090, São Paulo, SP, Brazil article info abstract Article history: A phylogenetic systematic perspective is instrumental in recovering new species and their evolutionary Received 8 December 2015 relationships. The advent of new technologies for molecular and morphological data acquisition and anal- Revised 7 April 2016 ysis, allied to the integration of knowledge from different areas, such as ecology and population genetics, Accepted 11 July 2016 allows for the emergence of more rigorous, accurate and complete scientific hypothesis on species diver- Available online xxxx sity. -

Activity of the Insectivorous Bat Pteronotus Parnellii Relative to Insect Resources and Vegetation Structure
Journal of Mammalogy, 96(5):1036–1044, 2015 DOI:10.1093/jmammal/gyv108 Activity of the insectivorous bat Pteronotus parnellii relative to insect resources and vegetation structure Leonardo Queiroz de Oliveira, Rodrigo Marciente, William E. Magnusson, and Paulo Estefano D. Bobrowiec* Coordenação de Biodiversidade, Instituto Nacional de Pesquisas da Amazônia (INPA), Av. André Araújo 2936, CP 2223, Manaus-AM 69080-971, Brazil Downloaded from Downloaded from by guest 2020 May on 19 https://academic.oup.com/jmammal/article-abstract/96/5/1036/917312 * Correspondent: [email protected] Riparian areas oten are assumed to be necessary sites or oraging by insectivorous bats because o high insect availability and ease o movement and echolocation in the orest. However, eects o vegetation clutter and insect availability on bat activity have not been compared between riparian and nonriparian areas. We used autonomous recorders to evaluate the eects o vegetation structure, insect mass, and assemblage composition on the activity o the aerial insectivorous bat Pteronotus parnellii along stream channels and nonriparian areas in a tropical rainorest in central Brazilian Amazonia. We quantied vegetation clutter using horizontal photographs, captured nocturnal insects with light traps, and recorded bat activity or 110 nights (1,320 h) in 22 sampling plots. Pteronotus parnellii was more active in sites with dense understory vegetation, which were more common away rom riparian zones. Bat activity was related to insect availability (mass and composition), independent o the habitat type. Ability to detect insects on vegetation and avoid obstacles should not restrict the activity o P. parnellii in cluttered sites. This suggests that mass and species composition o insects had stronger infuences on habitat use than did vegetation clutter. -

Chiroptera: Mormoopidae) in Ecuador; Implications for Its Conservation Therya, Vol
Therya E-ISSN: 2007-3364 [email protected] Asociación Mexicana de Mastozoología México CAMACHO, M. ALEJANDRA; LEIVA-D, VERÓNICA; LÓPEZ-WILCHIS, RICARDO; BURNEO, SANTIAGO F. Genetic diversity of the ghost-faced bat Mormoops megalophylla Peters, 1864 (Chiroptera: Mormoopidae) in Ecuador; implications for its conservation Therya, vol. 8, núm. 3, 2017, pp. 223-231 Asociación Mexicana de Mastozoología Baja California Sur, México Available in: http://www.redalyc.org/articulo.oa?id=402352772007 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative THERYA, 2017, Vol. 8 (3): 223-232 DOI: 10.12933/therya- 17-508 ISSN 2007-3364 Genetic diversity of the ghost-faced bat Mormoops megalophylla Peters, 1864 (Chiroptera: Mormoopidae) in Ecuador; implications for its conservation M. A LEJ ANDR A C AMAC HO 1* ,VERÓNI CA LEI VA -D.1, RICA RDO LÓPEZ -W IL CHIS 2 AND SANTI AGO F. B URNEO 1 1 Sección Mastozoología, Museo de Zoología, Escuela de Ciencias Biológicas, Facultad de Ciencias Exactas y Naturales, Ponticia Universidad Católica del Ecuador. Av. 12 de Octubre 1076 y Roca, 170525, Quito, Ecuador. E-mail: [email protected] (MAC), [email protected] (VLD), [email protected] (SFB). 2 Laboratorio de Biología y Ecología de Mamíferos, Departamento de Biología, Universidad Autónoma Metropolitana, Unidad Iztapalapa, Av. San Rafael Atlixco No. 186, Col. Vicentina, 09340. Ciudad de México, México. E-mail: [email protected] (RLW). * Corresponding author Mormoops megalophylla is a cave-dwelling bat distributed from southern United States across Central America to northern Peru. -

Phylogenetic Relationships of Mormoopid Bats (Chiroptera: Mormoopidae) Based on Morphological Data
amnb 00189 Mp 1 File # 01TQ PHYLOGENETIC RELATIONSHIPS OF MORMOOPID BATS (CHIROPTERA: MORMOOPIDAE) BASED ON MORPHOLOGICAL DATA NANCY B. SIMMONS Associate Curator Division of Vertebrate Zoology (Mammalogy) American Museum of Natural History TENLEY M. CONWAY Scientific Assistant Division of Vertebrate Zoology (Mammalogy) American Museum of Natural History BULLETIN OF THE AMERICAN MUSEUM OF NATURAL HISTORY Number 258, 97 pp., 12 figures, 4 tables Issued February 15, 2001 Price: $10.10 a copy Copyright ᭧ American Museum of Natural History 2001 ISSN 0003-0090 2 BULLETIN AMERICAN MUSEUM OF NATURAL HISTORY NO. 258 CONTENTS Abstract ....................................................................... 3 Introduction .................................................................... 3 Historical Background ......................................................... 3 Mormoopid Species: A Synopsis ............................................... 8 Goals of the Present Study .................................................... 12 Materials and Methods ......................................................... 12 Taxonomic Sampling, Outgroups, and Tree Rooting .............................. 12 Sources of Data ............................................................. 13 Definition of Characters and Ordering of Character States ........................ 13 Polarity ..................................................................... 15 Completeness ............................................................... 15 Methods of Phylogenetic Analysis -

Order Insectivora - Insectivores Family Soricidae- Shrews
Checklist of the Mammals of the Chiricahua Region, Cochise County, Arizona (1978) By E. Lendell Cockrum and Yar Petryszyn Department of Ecology and Evolutionary Biology University of Arizona (Taxonomy and common names as given J. Knox Jones et. al.in “Revised Checklist of North American Mammals North of Mexico, “ Occasional Papers, The Museum, Texas Tech University, no. 28, p. 1-14, 7 March 1975. To this has been added the recently named Sorex arizonae, Diersing and Hoffmeister.) An asterisk (*) indicates the species seen yet not recently collected in the region. Order Insectivora - Insectivores Family Soricidae- Shrews Vagrant Shrew Sorex vagrans Arizona Shrew Sorex arizonae Desert Shrew Notiosorex crawfordi Order Chiroptera – Bats Family Mormoopidae – Mormoopid Bats *Ghost-faced Bat Mormoops megalophylla Family Phyllostomatidae – Phyllostomatid Bats Long-tongued Bat Choeronycteris mexicana Mexican Long-nosed Bat Leptonycteris nivalis Sanborn’s Long-nosed Bat Leptonycteris sanborni Family Vespertilionidae – Vespertilionid Bats Yuma Myotis Myotis yumanensis Cave Myotis Myotis velifer Southwestern Myotis Myotis auriculus Fringed Myotis Myotis thysanodes Long-legged Myotis Myotis volans California Myotis Myotis californicus Small-footed Myotis Myotis leibii Silver-haired Bat Lasionycteris noctivagans Western Pipistrelle Pipistrellus hesperus Big Brown Bat Eptesicus fuscus Red Bat Lasiurus borealis Hoary Bat Lasiurus cinereus Southern Yellow Bat Lasiurus ega *Spotted Bat Euderma maculatum Townsend’s Big-eared Bat Plecotus townsendii Allen’s