Mucin Utilisation and Host Interactions of the Novel Intestinal
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prokaryotes (Domains Bacteria & Archaea)
2/4/15 Prokaryotes (Domains Bacteria & Archaea) KEY POINTS 1. Decomposers: recycle organic and inorganic molecules in environment; makes them available to other organisms. 2. Essential components of symbioses. 3. Encompasses the origins of metabolism and metabolic diversity. 4. Origin of photosynthesis and formation of atmospheric Oxygen Ceno- Meso- zoic zoic ANTIQUITY Humans Paleozoic Colonization of land Animals Origin of solar system and Earth • >3.5 BILLION years old. • Alone for 2 1 4 billion years Proterozoic Archaean Prokaryotes Billions of 2 years ago3 Multicellular eukaryotes Single-celled eukaryotes Atmospheric oxygen General characteristics 1. Small: compare to 10-100µm for 0.5-5µm eukaryotic cell; single-celled; may form colonies. 2. Lack membrane- enclosed organelles. 3. Cell wall present, but different from plant cell wall. 1 2/4/15 General characteristics 4. Occur everywhere, most numerous organisms. – More individuals in a handful of soil then there are people that have ever lived. – By far more individuals in our gut than eukaryotic cells that are actually us. General characteristics 5. Metabolic diversity established nutritional modes of eukaryotes. General characteristics 6. Important decomposers and recyclers 2 2/4/15 General characteristics 6. Important decomposers and recyclers • Form the basis of global nutrient cycles. General characteristics 7. Symbionts!!!!!!! • Parasites • Pathogenic organisms. • About 1/2 of all human diseases are caused by Bacteria General characteristics 7. Symbionts!!!!!!! • Parasites • Pathogenic organisms. • Extremely important in agriculture as well. Pierce’s disease is caused by Xylella fastidiosa, a Gamma Proteobacteria. It causes over $56 million in damage annually in California. That’s with $34 million spent to control it! = $90 million in California alone. -

Gastrointestinal Status and Microbiota Shaping in Amyotrophic Lateral Sclerosis: a New Frontier for Targeting?
8 Gastrointestinal Status and Microbiota Shaping in Amyotrophic Lateral Sclerosis: A New Frontier for Targeting? Letizia Mazzini1 • Fabiola De Marchi1 • Elena Niccolai2 • Jessica Mandrioli3, Amedeo Amedei2 1ALS Centre, Department of Neurology, Maggiore della Carità Hospital, University of Piemonte Orientale, Novara, Italy; 2Department of Experimental and clinical Medicine, University of Florence, Firenze, Italy; 3Department of Biomedical, Metabolic and Neural Science, University of Modena and Reggio Emilia, Modena, Italy Author for correspondence: Letizia Mazzini, ALS Centre, Department of Neurology, Maggiore della Carità Hospital, University of Piemonte Orientale, Novara, Italy. Email: [email protected] Doi: https://doi.org/10.36255/exonpublications.amyotrophiclateralsclerosis. microbiota.2021 Abstract: Amyotrophic lateral sclerosis (ALS) is a rare and severe neurodegenera- tive disease affecting the upper and lower motor neurons, causing diffuse muscle paralysis. Etiology and pathogenesis remain largely unclear, but several environ- mental, genetic, and molecular factors are thought to be involved in the disease process. Emerging data identify a relationship between gut microbiota dysbiosis and neurodegenerative diseases, such as Parkinson’s disease, Alzheimer’s disease, and ALS. In these disorders, neuroinflammation is being increasingly recognized as a driver for disease onset and progression. Gut bacteria play a crucial role in In: Amyotrophic Lateral Sclerosis. Araki T (Editor), Exon Publications, Brisbane, Australia. ISBN: -

A Generic Large-Scale Cause for Platelet Dysfunction and Depletion in Infection
Published online: 2020-04-12 THIEME 302 A Champion of Host Defense: A Generic Large-Scale Cause for Platelet Dysfunction and Depletion in Infection Martin J. Page, BSc (Hons)1 Etheresia Pretorius, PhD1 1 Department of Physiological Sciences, Stellenbosch University, Address for correspondence Etheresia Pretorius, PhD, Department of Stellenbosch, South Africa Physiological Sciences, Faculty of Science, Stellenbosch University, Private Bag X1 Matieland, Stellenbosch, 7602, South Africa Semin Thromb Hemost 2020;46:302–319. (e-mail: [email protected]). Abstract Thrombocytopenia is commonly associated with sepsis and infections, which in turn are characterized by a profound immune reaction to the invading pathogen. Platelets are one of the cellular entities that exert considerable immune, antibacterial, and antiviral actions, and are therefore active participants in the host response. Platelets are sensitive to surrounding inflammatory stimuli and contribute to the immune response by multiple mechanisms, including endowing the endothelium with a proinflammatory phenotype, enhancing and amplifying leukocyte recruitment and inflammation, promoting the effector functions of immune cells, and ensuring an optimal adaptive immune response. During infection, pathogens and their products influence the platelet response and can even be toxic. However, platelets are able to sense and engage bacteria and viruses to assist in their removal and destruction. Platelets greatly contribute to host defense by multiple mechanisms, including forming immune complexes and aggregates, shedding their granular content, and internalizing pathogens and subsequently being marked for removal. These processes, and the nature of platelet function in general, cause the platelet to be irreversibly consumed in Keywords the execution of its duty. An exaggerated systemic inflammatory response to infection ► platelets can drive platelet dysfunction, where platelets are inappropriately activated and face ► virus immunological destruction. -

Characterization of Α-L-Fucosidase and Other Digestive Hydrolases From
Acta Tropica 141 (2015) 118–127 Contents lists available at ScienceDirect Acta Tropica journal homepage: www.elsevier.com/locate/actatropica Characterization of ␣-L-fucosidase and other digestive hydrolases from Biomphalaria glabrata Natalia N. Perrella a,b, Rebeca S. Cantinha c,d, Eliana Nakano c, Adriana R. Lopes a,∗ a Laboratory of Biochemistry and Biophysics—Instituto Butantan, São Paulo, Brazil b Programa de Pós Graduac¸ ão Interunidades em Biotecnologia PPIB, Universidade de São Paulo, São Paulo, SP, Brazil c Laboratory of Parasitology—Instituto Butantan, São Paulo, Brazil d Instituto de Pesquisas Energéticas e Nucleares, Universidade de São Paulo, São Paulo, SP, Brazil article info abstract Article history: Schistosoma mansoni is one of the major agents of the disease Schistosomiasis, which is one of the Received 10 February 2014 major global public health concerns. Biomphalaria glabrata is an obligate intermediate mollusc host of Received in revised form 3 July 2014 S. mansoni. Although the development of S. mansoni occurs in the snail hepatopancreas, studies that Accepted 12 August 2014 focus on this organ remain limited. In this study, we biochemically identified five distinct carbohy- Available online 16 September 2014 drases (amylase, maltase, ␣-glucosidase, trehalase, and ␣-L-fucosidase), lipases, and peptidases in the B. glabrata hepatopancreas and focused on the isolation and characterization of the activity of ␣-L- Keywords: fucosidase. The isolated ␣-L-fucosidase has a molecular mass of 141 kDa, an optimum pH of 5.8, and Hepatopancreas ␣ Enzymes is inhibited by Tris, fucose, and 1-deoxyfuconojirimycin. B. glabrata -L-fucosidase is an exoglycosidase ␣-L-Fucosidase that can hydrolyze the natural substrate fucoidan to fucose residues. -

Glycoproteomics-Based Signatures for Tumor Subtyping and Clinical Outcome Prediction of High-Grade Serous Ovarian Cancer
ARTICLE https://doi.org/10.1038/s41467-020-19976-3 OPEN Glycoproteomics-based signatures for tumor subtyping and clinical outcome prediction of high-grade serous ovarian cancer Jianbo Pan 1,2,3, Yingwei Hu1,3, Shisheng Sun 1,3, Lijun Chen1, Michael Schnaubelt1, David Clark1, ✉ Minghui Ao1, Zhen Zhang1, Daniel Chan1, Jiang Qian2 & Hui Zhang 1 1234567890():,; Inter-tumor heterogeneity is a result of genomic, transcriptional, translational, and post- translational molecular features. To investigate the roles of protein glycosylation in the heterogeneity of high-grade serous ovarian carcinoma (HGSC), we perform mass spectrometry-based glycoproteomic characterization of 119 TCGA HGSC tissues. Cluster analysis of intact glycoproteomic profiles delineates 3 major tumor clusters and 5 groups of intact glycopeptides. It also shows a strong relationship between N-glycan structures and tumor molecular subtypes, one example of which being the association of fucosylation with mesenchymal subtype. Further survival analysis reveals that intact glycopeptide signatures of mesenchymal subtype are associated with a poor clinical outcome of HGSC. In addition, we study the expression of mRNAs, proteins, glycosites, and intact glycopeptides, as well as the expression levels of glycosylation enzymes involved in glycoprotein biosynthesis pathways in each tumor. The results show that glycoprotein levels are mainly controlled by the expression of their individual proteins, and, furthermore, that the glycoprotein-modifying glycans cor- respond to the protein levels of glycosylation enzymes. The variation in glycan types further shows coordination to the tumor heterogeneity. Deeper understanding of the glycosylation process and glycosylation production in different subtypes of HGSC may provide important clues for precision medicine and tumor-targeted therapy. -

Killing of Gram-Negative Bacteria by Lactoferrin and Lysozyme
Killing of gram-negative bacteria by lactoferrin and lysozyme. R T Ellison 3rd, T J Giehl J Clin Invest. 1991;88(4):1080-1091. https://doi.org/10.1172/JCI115407. Research Article Although lactoferrin has antimicrobial activity, its mechanism of action is not full defined. Recently we have shown that the protein alters the Gram-negative outer membrane. As this membrane protects Gram-negative cells from lysozyme, we have studied whether lactoferrin's membrane effect could enhance the antibacterial activity of lysozyme. We have found that while each protein alone is bacteriostatic, together they can be bactericidal for strains of V. cholerae, S. typhimurium, and E. coli. The bactericidal effect is dose dependent, blocked by iron saturation of lactoferrin, and inhibited by high calcium levels, although lactoferrin does not chelate calcium. Using differing media, the effect of lactoferrin and lysozyme can be partially or completely inhibited; the degree of inhibition correlating with media osmolarity. Transmission electron microscopy shows that E. coli cells exposed to lactoferrin and lysozyme at 40 mOsm become enlarged and hypodense, suggesting killing through osmotic damage. Dialysis chamber studies indicate that bacterial killing requires direct contact with lactoferrin, and work with purified LPS suggests that this relates to direct LPS-binding by the protein. As lactoferrin and lysozyme are present together in high levels in mucosal secretions and neutrophil granules, it is probable that their interaction contributes to host defense. Find the latest version: https://jci.me/115407/pdf Killing of Gram-negative Bacteria by Lactofernn and Lysozyme Richard T. Ellison III*" and Theodore J. Giehl *Medical and tResearch Services, Department of Veterans Affairs Medical Center, and Division ofInfectious Diseases, Department ofMedicine, University ofColorado School ofMedicine, Denver, Colorado 80220 Abstract (4). -

Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer
cancers Article Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer Jin-Yong Jeong 1, Tae-Bum Kim 2 , Jinju Kim 1, Hwi Wan Choi 1, Eo Jin Kim 1, Hyun Ju Yoo 1 , Song Lee 3, Hye Ryeong Jun 3, Wonbeak Yoo 4 , Seokho Kim 5, Song Cheol Kim 3,6,* and Eunsung Jun 1,3,* 1 Department of Convergence Medicine, Asan Institute for Life Sciences, University of Ulsan College of Medicine and Asan Medical Center, Seoul 05505, Korea; [email protected] (J.-Y.J.); [email protected] (J.K.); [email protected] (H.W.C.); [email protected] (E.J.K.); [email protected] (H.J.Y.) 2 Department of Allergy and Clinical Immunology, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, Korea; [email protected] 3 Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, Asan Medical Center, University of Ulsan College of Medicine, Seoul 05505, Korea; [email protected] (S.L.); [email protected] (H.R.J.) 4 Environmental Disease Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon 34141, Korea; [email protected] 5 Department of Medicinal Biotechnology, College of Health Sciences, Dong-A University, Busan 49315, Korea; [email protected] 6 Biomedical Engineering Research Center, Asan Institute of Life Science, AMIST, Asan Medical Center, Seoul 05505, Korea * Correspondence: [email protected] (S.C.K.); [email protected] (E.J.); Tel.: +82-2-3010-3936 (S.C.K.); +82-2-3010-1696 (E.J.); Fax: +82-2-474-9027 (S.C.K.); +82-2-474-9027 (E.J.) Received: 13 July 2020; Accepted: 17 August 2020; Published: 19 August 2020 Abstract: This study was conducted to identify the composition and diversity of the microbiome in tissues of pancreatic cancer and to determine its role. -
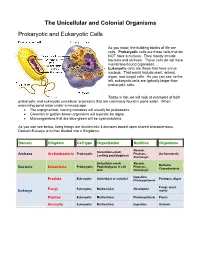
The Unicellular and Colonial Organisms Prokaryotic And
The Unicellular and Colonial Organisms Prokaryotic and Eukaryotic Cells As you know, the building blocks of life are cells. Prokaryotic cells are those cells that do NOT have a nucleus. They mostly include bacteria and archaea. These cells do not have membrane-bound organelles. Eukaryotic cells are those that have a true nucleus. That would include plant, animal, algae, and fungal cells. As you can see, to the left, eukaryotic cells are typically larger than prokaryotic cells. Today in lab, we will look at examples of both prokaryotic and eukaryotic unicellular organisms that are commonly found in pond water. When examining pond water under a microscope… The unpigmented, moving microbes will usually be protozoans. Greenish or golden-brown organisms will typically be algae. Microorganisms that are blue-green will be cyanobacteria. As you can see below, living things are divided into 3 domains based upon shared characteristics. Domain Eukarya is further divided into 4 Kingdoms. Domain Kingdom Cell type Organization Nutrition Organisms Absorb, Unicellular-small; Prokaryotic Photsyn., Archaeacteria Archaea Archaebacteria Lacking peptidoglycan Chemosyn. Unicellular-small; Absorb, Bacteria, Prokaryotic Peptidoglycan in cell Photsyn., Bacteria Eubacteria Cyanobacteria wall Chemosyn. Ingestion, Eukaryotic Unicellular or colonial Protozoa, Algae Protista Photosynthesis Fungi, yeast, Fungi Eukaryotic Multicellular Absorption Eukarya molds Plantae Eukaryotic Multicellular Photosynthesis Plants Animalia Eukaryotic Multicellular Ingestion Animals Prokaryotic Organisms – the archaea, non-photosynthetic bacteria, and cyanobacteria Archaea - Microorganisms that resemble bacteria, but are different from them in certain aspects. Archaea cell walls do not include the macromolecule peptidoglycan, which is always found in the cell walls of bacteria. Archaea usually live in extreme, often very hot or salty environments, such as hot mineral springs or deep-sea hydrothermal vents. -

Characterization of the First Cultured Representative of Verrucomicrobia Subdivision 5 Indicates the Proposal of a Novel Phylum
The ISME Journal (2016) 10, 2801–2816 OPEN © 2016 International Society for Microbial Ecology All rights reserved 1751-7362/16 www.nature.com/ismej ORIGINAL ARTICLE Characterization of the first cultured representative of Verrucomicrobia subdivision 5 indicates the proposal of a novel phylum Stefan Spring1, Boyke Bunk2, Cathrin Spröer3, Peter Schumann3, Manfred Rohde4, Brian J Tindall1 and Hans-Peter Klenk1,5 1Department Microorganisms, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany; 2Department Microbial Ecology and Diversity Research, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany; 3Department Central Services, Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany and 4Central Facility for Microscopy, Helmholtz-Centre of Infection Research, Braunschweig, Germany The recently isolated strain L21-Fru-ABT represents moderately halophilic, obligately anaerobic and saccharolytic bacteria that thrive in the suboxic transition zones of hypersaline microbial mats. Phylogenetic analyses based on 16S rRNA genes, RpoB proteins and gene content indicated that strain L21-Fru-ABT represents a novel species and genus affiliated with a distinct phylum-level lineage originally designated Verrucomicrobia subdivision 5. A survey of environmental 16S rRNA gene sequences revealed that members of this newly recognized phylum are wide-spread and ecologically important in various anoxic environments ranging from hypersaline sediments to wastewater and the intestine of animals. Characteristic phenotypic traits of the novel strain included the formation of extracellular polymeric substances, a Gram-negative cell wall containing peptidoglycan and the absence of odd-numbered cellular fatty acids. Unusual metabolic features deduced from analysis of the genome sequence were the production of sucrose as osmoprotectant, an atypical glycolytic pathway lacking pyruvate kinase and the synthesis of isoprenoids via mevalonate. -

Akkermansia Muciniphila Ameliorates the Age-Related Decline in Colonic
van der Lugt et al. Immunity & Ageing (2019) 16:6 https://doi.org/10.1186/s12979-019-0145-z RESEARCH Open Access Akkermansia muciniphila ameliorates the age-related decline in colonic mucus thickness and attenuates immune activation in accelerated aging Ercc1−/Δ7 mice Benthe van der Lugt1*†, Adriaan A. van Beek2,3†, Steven Aalvink4, Ben Meijer3, Bruno Sovran5, Wilbert P. Vermeij6,7, Renata M. C. Brandt6, Willem M. de Vos4,8, Huub F. J. Savelkoul3, Wilma T. Steegenga1 and Clara Belzer4* Abstract Background: The use of Akkermansia muciniphila as potential therapeutic intervention is receiving increasing attention. Health benefits attributed to this bacterium include an improvement of metabolic disorders and exerting anti-inflammatory effects. The abundance of A. muciniphila is associated with a healthy gut in early mid- and later life. However, the effects of A. muciniphila on a decline in intestinal health during the aging process are not investigated yet. We supplemented accelerated aging Ercc1−/Δ7 mice with A. muciniphila for 10 weeks and investigated histological, transcriptional and immunological aspects of intestinal health. Results: The thickness of the colonic mucus layer increased about 3-fold after long-term A. muciniphila supplementation and was even significantly thicker compared to mice supplemented with Lactobacillus plantarum WCFS1. Colonic gene expression profiles pointed towards a decreased expression of genes and pathways related to inflammation and immune function, and suggested a decreased presence of B cells in colon. Total B cell frequencies in spleen and mesenteric lymph nodes were not altered after A. muciniphila supplementation. Mature and immature B cell frequencies in bone marrow were increased, whereas B cell precursors were unaffected. -

Identification and Characterisation of Mucin Abnormalities in the Ganglionic Bowel in Hirschsprung’S Disease
IDENTIFICATION AND CHARACTERISATION OF MUCIN ABNORMALITIES IN THE GANGLIONIC BOWEL IN HIRSCHSPRUNG’S DISEASE By Ruth Marian Speare Submitted for MD (Res) 1 Abstract Hirschsprung’s disease (HD) is a congenital abnormality of unknown origin, characterised by a lack of ganglion cells in the distal colon which results in functional colonic obstruction. The major cause of morbidity and mortality in these children results from an inflammatory condition called enterocolitis. Mucins are large glycoproteins produced by intestinal cells which are a vital part of the colonic defensive barrier to infection. Previous work has found that this barrier is deficient in children with HD in the aganglionic colon and in the immediately adjacent ganglionic colon and that this is related to the risk of developing enterocolitis. This study aimed to further investigate the mucin defensive barrier in a greater region of the ganglionic colon in HD, to establish the extent of any mucin deficiencies and whether these were confined to a limited region close to the aganglionic colon. Mucosal biopsies were collected at intervals along the colon at the time of corrective surgery or colostomy closure in the controls. Organ culture with radioactive mucin precursors was performed and the mucin produced was purified and analysed, results quantified by DNA content in the sample. Lectin binding studies were also carried out. Patients were found to produce lower levels of new mucins most distally, but much higher levels five centimetres proximal when compared to controls. The rest of the colon studied also showed changes in mucin production, with a lack of production of gel-forming mucins and sulphated secreted mucins in Hirschsprung’s disease higher up the colon. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota.