Normal Brain
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

LES FACULTÉS DE DROIT FRANÇAISES AU Xvr SIÈCLE ÉLÉMENTS DE BIBLIOGRAPHIE (2E Partie)
LES FACULTÉS DE DROIT FRANÇAISES AU xvr SIÈCLE ÉLÉMENTS DE BIBLIOGRAPHIE (2e partie) Nous poursuivons la publication de la bibliographie de M. Jean Louis Thireau, Maître de conférences à l'Université de Paris XII (1). Faculté de droit de Cahors Articles • BAUDEL M.-J., MALINOWSKI J.-Ph., ((Histoire de l'Université de Cahors», Bull. Soc. Etudes litt., sci. artist. Lot, t. 2 (1875), p. 135-192, 288-320; t. 3 (1876), p. 201-240, 273-300; t. 4 (1878), p. 126-176. • BAUDEL M.-J., « François Roaldès, docteur régent de l'Université de Cahors (1513-1589), biographie», Bull. Soc. Etudes litt., sci, artist. Lot, t. 3 (1876), p. 190-200. • BRESSOLES G., « Un manuscrit de François Roaldès, professeur à l'Université de Cahors, sur le droit de fouage », Rec. Acad. Législ. Toulouse, t. 27 (1878), p. 302-314. ·• CAILLEMER E., « Etude sur Antoine de Govéa », Mém. Acad. impé riale Sei., Arts Belles Lettres Caen, 1865, p. 79-120. • SERRAO J.-V., «Antonio de Gouveia e o seu tempo», Boletim da Faculdade de Direito da Universidade de Coimbra, t. 42 (1966), p. 25-224; t. 43 (1967), p. 1-131. • « Texte d'un contrat passé en 1585 entre l'Université de Cahors et François Roaldès par lequel ce dernier s'engageait à professer pen dant neuf ans le droit civil et le droit canon dans cette Université», Rec. Acad. Législ. Toulouse, t. 10 (1861), p. 545-548. (1) V. la première partie, cette revue, 1987, n° 5, pp. 101 ss. 178 REVUE D'HISTOIRE DES FACULTÉS DE DROIT Faculté de droit de Dole Ouvrages • APPLETON Ch., Coup d'œil bibliographique sur deux jurisconsultes français du XVI• siècle. -
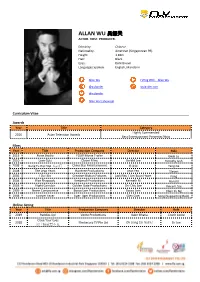
Allan Wu 吴振天 Actor
ALLAN WU 吴振天 ACTOR. HOST. PRODUCER. Ethnicity: Chinese Nationality: American (Singaporean PR) Height: 1.83m Hair: Black Eyes: Dark Brown Languages Spoken: English, Mandarin Allan Wu FLYing With... Allan Wu @wulander wulander.com @wulander Allan Wu's Showreel Curriculum Vitae Awards Year Title Category Highly Commended 2016 Asian Television Awards Best Entertainment Presenter/Host Films Year Title Production Company Director Role 2015 Poise Studio FOUR Movie Trailer Gwai Lo 2010 Love Cuts Clover Films Gerald Lee Timothy Goh 2008 Kung Fu Hip Hop 精武门 China Star Entertainment 傅华阳 Tang Ge 2008 The Leap Years Raintree Productions Jean Yeo Steven 2005 I Do I Do Creative Motion Pictures Jack Neo / Lim Boon Hwee Feng 2004 Rice Rhapsody Kenberoli Productions Kenneth Bi Ronald 2003 Night Corridor Golden Gate Productions Chi-Chiu Lee Vincent Sze 1999 Never Compromise Bosco Lam Productions Bosco Lam Chun Yu Ng 1998 Forever Fever Tiger Tiger Productions Glen Goei Seng (Supporting Role) Online Acting Year Title Production Company Director Role 2019 Paddles Up! Verite Productions Kabir Bhatia Coach Leslie Close Your Eyes 2018 Mediacorp TV Pte Ltd Oh Liang Cai 胡凉财 Dr Joe 闭上眼就看不见 Television Actor Year Title Production Company Director Role 2019 CLIF 5 Mediacorp TV Pte Ltd Oh Liang Cai Alexis 2018 Divided 分裂 Mediacorp TV Pte Ltd Lee Thean-jeen Inspector Wu Wenjie 2014 Mata Mata Season 2 MediaCorp TV Singapore Pte Ltd Alan Leong 2014 First Gear House Films Lead 2006 House Of Joy Mediacorp Studios Pte Ltd 罗温温 Zheng Sanji 2006 Xing Jing Er Ren Zhu Mediacorp -

Pierre Ayrault V. the Society of Jesus: Gallican-Robin Constructions of Paternal Authority on Legal, Political, and Religious Grounds
Pierre Ayrault v. The Society of Jesus: Gallican-robin Constructions of Paternal Authority on Legal, Political, and Religious Grounds Katherine Godwin A thesis submitted in partial fulfillment of the requirements for the degree of BACHELOR OF ARTS WITH HONORS DEPARTMENT OF HISTORY UNIVERSITY OF MICHIGAN March 31, 2008 Advised by Professor Michael MacDonald and Professor George Hoffmann TABLE OF CONTENTS Acknowledgments………………………………………………………………………………ii. Introduction: Biography………………………………………………………………………..1. Chapter One: Une loi…………………………………………………………………………..17. Parlement vs. The Council of Trent……………………………………………………...22. Contumacy……………………………………………………………………………….30. Rapt....................................................................................................................................37. Life, Liberty, and Bourgeoisie…………………………………………………………...50. Chapter Two: Un roi…………………………………………………………………………...53. “This domestic monarchy”………………………………………………………………55. Seduction and Sedition…………………………………………………………………..63. Politiques………………………………………………………………………………...74. Chapter Three: Une foi………………………………………………………………………...84. Piety: The Ten Commandments and Reconciliation…………………………………….87. False Monks…………………………………………………………………………….104. Conclusion……………………………………………………………………………………..116. Bibliography…………………………………………………………………………………..122. Acknowledgements Of course, any project of this magnitude never comes to completion through the efforts of one single person. I would like to thank Professors Michael MacDonald and George Hoffmann for their generosity and -

Corporate Profile
Corporate Profile! 2018 Fast Growing ! F&B Distributor! Copyright Provenance®distributions 2 Who We Are! Provenance® Distributions Pte Ltd is a fast growing company that imports and distributes fast-moving food & beverages (F&B) in Singapore, Hong Kong & Macau. We also re-export F&B to other Asian markets. The South China Morning Post — Asia’s most widely read English newspaper based in Hong Kong — featured us as ”A Rising Star in Asia.” (August 9, 2017) & The Straits Times — Singapore’s leading newspaper — also featured us on Singaporean brand successes in Hong Kong (Sep 9, 2018). We have an extensive B2B distribution network — covering 1,700+ locations — and distribute global brands such as S&W fruit juices and leading Singaporean brands such as The Golden Duck. We are an Official Partner of MICHELIN Guide for their events in the region. Through our strong marketing expertise, we run impactful marketing campaigns which generate brand awareness and sales. We are also sales & CRM experts with a large database of B2B purchaser contacts. Our vision is to become a leading distributor in Asia. Copyright Provenance®distributions 3 Rising Star in Asia! *As reported in the South China Morning Post, Business Section, 9th Aug 2017 Copyright Provenance®distributions 4 Freight Sales Marketing Suite of Services! Delivery Copyright Provenance®distributions Warehousing Orders Merchandising Receivables 5 Our Offices! China (Shenzhen) (In 2019) Singapore HQ Hong Kong Macau 318 Tanglin Rd Two IFC Praia Grand Copyright Provenance®distributions 6 Featured in -

C Ntentasia 19 Sept-2 Oct 2016 19 September-2 October 2016 Page 2
19 September- 2 October 2016 ! s r a ye 2 0 C 016 g 1 NTENT - Celebratin www.contentasia.tv l www.contentasiasummit.com APOS l Tech Turner’s Tuzki enters China’s movie space debuts in China Feature film under way with Tencent Pictures MAIN COLOR PALETTE 10 GRADIENT BG GRADIENT R: 190 G: 214 B: 48 Focus onR: 0 tech’sG: 0 B: 0 role in Take the green and the blue Take the green and the blue C: 30 M: 0 Y: 100 K: 0 C: 75 M: 68 Y: 67 K: 90 from the main palette. from the main palette. Opacity: 100% Opacity: 50% R: 0 G: 80 B: 255 unlockingR: 138 G: 140value B: 143 from Blending Mode: Normal Blending Mode: Hue C: 84 M: 68 Y: 0 K: 0 C: 49 M: 39 Y: 38 K: 3 consumer eyeballs/wallets Media Partners Asia (MPA) debuts APOS l Tech on Tuesday evening, setting hard-core tech/broadband/distribution conversations against the backdrop of one of China’s most scenic locations. MPA says media and telecom opera- tors are “at a critical crossroads amidst changes in CPE technology, the evolu- tion of broadband, technology con- solidation and the emergence of onine video”. The question, says executive director Vivek Couto, “is how operators are gear- ing to address these challenges with new and existing products”. The rest of the story is on page 4 Tuzki LeSports, NBA in Turner Asia Pacific has entered mainland centres. HK, Macau tie-up China’s movie rush with a feature film The MoU was announced out of Beijing based on the 10-year-old Tuzki emoticon. -

2015-Sgipos-14.Pdf
IN THE HEARINGS AND MEDIATION GROUP OF THE INTELLECTUAL PROPERTY OFFICE OF SINGAPORE REPUBLIC OF SINGAPORE Trade Mark No T1114593Z and T1114594H Hearing Date: 29 April 2015 (Further Written Submissions: 26 May 2015) IN THE MATTER OF A TRADE MARK APPLICATION BY DOCHIRNIE PIDPRYIEMSTVO "KONDYTERSKA KORPORATSIIA "ROSHEN" AND OPPOSITION THERETO BY FERRERO S.P.A. Hearing Officer: Ms Diyanah Binte Baharudin Assistant Registrar of Trade Marks China Intellectual Property Agency for the Applicants (unrepresented at hearing) Mr Sukumar Karuppiah (Ravindran Associates) for the Opponents Cur Adv Vult GROUNDS OF DECISION 1 Dochirnie pidpryiemstvo "Kondyterska korporatsiia "Roshen" (the " Applicants ") applied to protect the following signs: Trade Mark Representation Class Specification Application No. T1114593Z 29 Meat; Fish, not live; Poultry, not live; Game, not live; Meat extracts; (referred to as the Preserved, frozen, dried and cooked "Applicants' fruits and vegetables; Eggs; Milk and ROSHEN milk products; Edible oils and fats; Mark ") Fruit jellies; Jams; Compotes. 30 Confectionery; Sugar confectionery; Sweetmeats [candy]; Candy for food; Fondants [confectionery]; Fruit jellies [confectionery]; Fruit Jellies [confectionery]; Peanut confectionery; Peppermint sweets; Chocolate; Coffee; Cookies; Pastry; bread; tea; cocoa; Coffee-based beverages; Coffee beverages with milk; Honey. T1114594H 29 Meat; Fish, not live; Poultry, not live; Game, not live; Meat extracts; (referred to as the Preserved, frozen, dried and cooked "Applicants' fruits and vegetables; Eggs; Milk and ROSHEN milk products; Edible oils and fats; CLASSIC Fruit jellies; Jams; Compotes. Mark ") 30 Confectionery; Sugar confectionery; Sweetmeats [candy]; Candy for food; Fondants [confectionery]; Fruit jellies [confectionery]; Fruit Jellies [confectionery]; Peanut confectionery; Peppermint sweets; Chocolate; Coffee; Cookies; Pastry; bread; tea; cocoa; Coffee-based beverages; Coffee beverages with milk; Honey. -

History and Religion Religionsgeschichtliche Versuche Und Vorarbeiten
History and Religion Religionsgeschichtliche Versuche und Vorarbeiten Herausgegeben von Jörg Rüpke und Christoph Uehlinger Band 68 History and Religion Narrating a Religious Past Edited by Bernd-Christian Otto, Susanne Rau and Jörg Rüpke with the support of Andrés Quero-Sánchez ISBN 978-3-11-044454-4 e-ISBN (PDF) 978-3-11-044595-4 e-ISBN (EPUB) 978-3-11-043725-6 ISSN 0939-2580 Library of Congress Cataloging-in-Publication Data A CIP catalog record for this book has been applied for at the Library of Congress. Bibliographic information published by the Deutsche Nationalbibliothek The Deutsche Nationalbibliothek lists this publication in the Deutsche Nationalbibliografie; detailed bibliographic data are available on the Internet at http://dnb.dnb.de. © 2015 Walter de Gruyter GmbH, Berlin/Boston Printing and binding: CPI books GmbH, Leck ∞ Printed on acid-free paper Printed in Germany www.degruyter.com TableofContents Historyand Religion 1 Section I Origins and developments Introduction 21 Johannes Bronkhorst The historiography of Brahmanism 27 Jörg Rüpke Construing ‘religion’ by doinghistoriography: The historicisation of religion in the Roman Republic 45 Anders Klostergaard Petersen The use of historiography in Paul: Acase-study of the instrumentalisation of the past in the context of Late Second Temple Judaism 63 Ingvild Sælid Gilhus Flirty fishing and poisonous serpents: Epiphanius of Salamis inside his Medical chestagainstheresies 93 Sylvie Hureau Reading sutras in biographies of Chinese Buddhist monks 109 Chase F. Robinson Historyand -

SELECTED WORKS of JOHN CALVIN VOL. 6 LETTERS 1554-1558 by John Calvin
THE AGES DIGITAL LIBRARY HISTORY SELECTED WORKS OF JOHN CALVIN VOL. 6 LETTERS 1554-1558 by John Calvin B o o k s F o r Th e A g e s AGES Software • Albany, OR USA Version 1.0 © 1998 2 SELECTED WORKS OF JOHN CALVIN TRACTS AND LETTERS EDITED BY HENRY BEVERIDGE AND JULES BONNET VOLUME 6 LETTERS, PART 3 1554-1558 3 CONTENTS 1554 LETTER 340 — TO VIRET. — Consolations and encouragements — election of the New Syndics at Geneva. LETTER 341 — TO AMBROSE BLAURER. — Friendly complaints respecting the silence of Blaurer — despatch of several writings. LETTER 342 — TO BULLINGER. — Reconciliation of parties — apparent tranquillity of the republic — announces the book against the errors of Servetus. LETTER 343 — TO A SEIGNEUR OF PIEDMONT. — He exhorts him to perseverance in the faith, in giving his children a christian education. LETTER 344 — TO THE BRETHREN OF ORBE. — Vows and counsels for the establishment of religious unity in their city by the abolition of the Catholic worship. LETTER 345 — TO VIRET. — Recommendation of several English and Scotch refugees. LETTER 346 — TO THE BRETHREN OF WEZEL. — Entreaty not to break the unity of the Church because of some diversities in the ceremonies. LETTER 347 — TO BULLINGER. — Recommendation of two English refugees — state of parties at Geneva — fresh persecutions in France — military movements of Henry II. — thanks for a work sent him — publication at Bale of a pamphlet on the repression of heresy by the sword of the magistrate — divers salutations. LETTER 348 — TO BULLINGER. — Explanations respecting the book against the errors of Servetus — answers to three questions of Knox. -

Voltaire and the Jura Serfs, 1770-1778
VOLTAI HE AND THE JURA SERFS by JOHN P.H. COLLINS, B.A~, B.Ed. A Thesis Submitted to the School of Graduate Studies in Partial Fu1fi1ment of the Requirements------------ for the Degree Master of Arts McMaster University August 1978 MASTER OF ARTS (1978) McMASTER UNIVERSITY Hamilton, Ontario TITLE: Voltaire and the Jura Serfs, 1770-1778 AUTHOR: John P.H. Collins, B.A. (McMaster University) B.Ed. (University of Western Ontario) SUPERVISOR: Dr. Pierre-Marie Conlon NUMBER OF PAGES: ix,133 ii ABSTRACT From 1770 until his death in 1778, Voltaire led a most vociferous campaign against the rernnants of feudalism in France. The emphasis of his campaign was placed on the institution of serfdom, a system of seigniorial rights which entitled a lord to specifie services and fees from his vassals. Voltaire's interest in serfdom was sparked by the fact that there existed sorne twelve thousand peasants living as serfs due to rights existing since the middle ages, at Saint-Claude, only a few miles from his estate of Ferney near the Swiss border. Voltaire's concern for these serfs was augmented further by the knowledge that the lords of Saint-Clauàe were in fact a group of twenty Benedictine monks. The aim of this dissertation is to examine Voltaire's campaign, not only for the emancipation of the -:'se-rfs-or SairiE-Claude-;-but.-Ior-tne -ano1:lt:ion--oÎ--- feudalism throughout France and for the establishment of a uniform code of law. This subject has never been fully investigated, and consequently, in light of the approach of the bicentennial anniversary of Voltaire's death, a study of the campaign for the serfs of the Jura mountains does seem appropriate. -

Chinese Face/Off
Chinese Face/Off THE TRANSNATIONAL POPULAR CULTURE OF HONG KONG KWAI-CHEUNG LO UNIVERSITY OF ILLINOIS PRESS Urbana and Chicago © 2005 by the Board of Trustees of the University of Illinois All rights reserved Manufactured in the United States of America 1 2 3 4 5 c p 5 4 3 2 1 6l This book is printed on acid-free paper. First published in 2005, this title is available from the University of Illinois Press except in Asia, Australia, and New Zealand, where it is available from Hong Kong University Press. The University of Illinois Press Hong Kong University Press 1325 South Oak Street 14/F Hing Wai Centre Champaign, IL 61820-6903 7 Tin Wan Praya Road www.press.uillinois.edu Aberdeen, Hong Kong ISBN 0-252-02978-x (cloth) www.hkupress.org 0-252-07228-6 (paper) ISBN 962-209-753-7 Library of Congress Cataloging-in-Publication Data Lo, Kwai-Cheung Chinese face/off: the transnational popular culture of Hong Kong I Kwai-Cheung Lo. p. cm. - (Popular culture and politics in Asia Pacific) Includes bibliographical references. ISBN 0-252-02978-x (cloth: alk. paper) - ISBN 0-252-07228-6 (pbk.: alk. paper) i. Popular culture-China-Hong Kong. 2. Hong Kong (China) Civilization-21st century. 3. Hong Kong (China)-Civilization Foreign influences. I. Title. II. Series. DS796.H75L557 2005 306'.095125-dc22 2004018122 Contents Acknowledgments vn Note on Transliteration IX Introduction: The Chineseness of Hong Kong's Transnational Culture in Today's World 1 Part 1: From Voice to Words and Back in Chinese Identification 1. -

Beyond Singapore Girl Anderson, B
Exploring the gendering of national subjects in Singapore The branding of Singapore International Airlines with the im- H Beyond age of a beautiful, petite and servile ‘Oriental’ woman dressed in U figure-huggingsarong-kebaya is one of the world’s longest running DS the and most successful advertising campaigns. But this image does not simply advertise a service; it is part of a global and national O Singapore Girl regime of symbolic constructions of gender that today is seen as N outdated and sexist, and bearing little relation to modern Singa- BEY Discourses of Gender and Nation in Singapore pore where women have good access to education and increased life choices resulting from engagement in the wage economy. The nation’s economic success has been a force for their liberation. O CHRIS HUDSON One catastrophic consequence of women’s changed lives has been T ND the plunge in fertility rates. Singapore has one of the world’s low- est despite energetic government campaigns encouraging women to have more babies – and men to be more ‘masculine’. The failure H of these campaigns and rethinking of the Singapore Girl highlight E S a key premise of this book: there are limits to the power of discur- sive constructions of gender in the national interest. I NGAP ABOUT THE AUTHOR Chris Hudson is a research leader in the Globalization and Culture Program at the Global Cities Research Institute as well as Associate OR Professor of Asian Media and Culture at RMIT University in Melbourne, Australia. E G IR L GENDERING ASIA a series on gender intersections www.niaspress.dk Hudson-pbk-cover.indd 1 01/08/2013 11:39 BEYOND THE SINGAPORE GIRL Hudson book.indd 1 01/08/2013 11:27 GENDERING ASIA A Series on Gender Intersections Gendering Asia is a well-established and exciting series addressing the ways in which power and constructions of gender, sex, sexuality and the body intersect with one another and pervade contemporary Asian societies. -

Reproductions Supplied by EDRS Are the Best That Can Be Made from the Original Document
DOCUMENT RESUME ED 439 312 CG 029 879 AUTHOR Evraiff, Bill, Comp.; Evraiff, Lois, Comp. TITLE Caring in an Age of Technology. Proceedings of the International Conference on Counseling in the 21st Century (6th, Beijing, China, May 29-30, 1997). PUB DATE 1997-05-00 NOTE 321p.; For individual papers, see CG 029 880-931. PUB TYPE Collected Wofks Proceedings (021) EDRS PRICE MF01/PC13 Plus Postage. DESCRIPTORS College Students; Computers; Counseling; *Cross Cultural Studies; Elementary Secondary Education; *Family Counseling; Foreign Countries; Foreign Culture; Higher Education; Internet; *Social Change; Technology ABSTRACT The two main themes of this conference were the influence of technology on families, and technology and counseling. Many of the papers consider the impact technology is having on individuals and families, and subsequently how it is affecting the counseling profession. This involves new ways of counseling using technological resources,and counseling techniques for concerns brought about by technology. Other topics covered include insights from mother-daughter groups, behavior counseling for parents with special children, human rights education, and career counseling. All presentations are included. The 52 papers include: (1) "The Homeroom Teacher's Role in Psychological Counseling at School"(W. Guodong); (2) "A Study of Family Therapy for Student Counseling" (0. Honda); and (3) "Technology and the School Counselor"(D. Coy and C. Minor). (JDM) Reproductions supplied by EDRS are the best that can be made from the original document. INCOUNSELING THE M-1 ST CENTURY BESTOOPYAVAILABLE U.S. DEPARTMENT OF EDUCATION Office of Educational Research and Improvement EDUCATIONAL RESOURCES INFORMATION "PERMISSION TO REPRODUCE THIS CENTER (ERIC) MATERIAL HAS BEEN GRANTED BY This document has been reproduced as received from the person or organization originating it.