Characterization of the S1 Subsite Specificity of Aspergillopepsin I by Site-Directed Mutagenesis1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
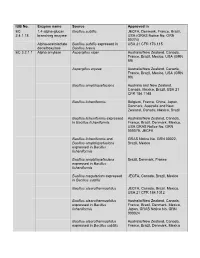
IUB No. Enzyme Name Source Approved in EC 1,4-Alpha-Glucan Bacillus Subtilis JECFA, Denmark, France, Brazil, 2.4.1.18 Branching Enzyme USA (GRAS Notice No
IUB No. Enzyme name Source Approved in EC 1,4-alpha-glucan Bacillus subtilis JECFA, Denmark, France, Brazil, 2.4.1.18 branching enzyme USA (GRAS Notice No. GRN 00274) Alpha-acetolactate Bacillus subtilis expressed in USA 21 CFR 173.115 decarboxylase Bacillus brevis EC 3.2.1.1 Alpha amylase Aspergillus niger Australia/New Zealand, Canada, France, Brazil, Mexico, USA (GRN 89) Aspergillus oryzae Australia/New Zealand, Canada, France, Brazil, Mexico, USA (GRN 90) Bacillus amyloliquefaciens Australia and New Zealand, Canada, Mexico, Brazil, USA 21 CFR 184.1148 Bacillus licheniformis Belgium, France, China, Japan, Denmark, Australia and New Zealand, Canada, Mexico, Brazil Bacillus licheniformis expressed Australia/New Zealand, Canada, in Bacillus licheniformis France, Brazil, Denmark, Mexico, USA GRAS Notice No. GRN 000079, JECFA Bacillus licheniformis and GRAS Notice No. GRN 00022, Bacillus amyloliquefaciens Brazil, Mexico expressed in Bacillus licheniformis Bacillus amyloliquefaciens Brazil, Denmark, France expressed in Bacillus licheniformis Bacillus megaterium expressed JECFA, Canada, Brazil, Mexico in Bacillus subtilis Bacillus stearothermophilus JECFA, Canada, Brazil, Mexico, USA 21 CFR 184.1012 Bacillus stearothermophilus Australia/New Zealand, Canada, expressed in Bacillus France, Brazil, Denmark, Mexico, licheniformis Japan, GRAS Notice No. GRN 000024 Bacillus stearothermophilus Australia/New Zealand, Canada, expressed in Bacillus subtilis France, Brazil, Denmark, Mexico Bacillus stearothermophilus Australia/New Zealand, Canada, (Geobacillus France, Europa, Brazil, Mexico, stearothermophilus) USA (GRN 594) Bacillus subtilis Australia and New Zealand, Canada, Mexico, Brazil, USA 21 CFR 184.1148 Rhizopus delemar Brazil Rhizopus oryzae Brazil, Mexico, USA GRAS Notice No. GRN 000090 Thermoccocales expressed in Brazil, USA (GRN126) Pseudomonas fluorecens Rhizomucor pusillus expressed Brazil, Denmark, France, Mexico, in Aspergillus niger Australia and New Zealand. -

(12) United States Patent (10) Patent No.: US 6,395,889 B1 Robison (45) Date of Patent: May 28, 2002
USOO6395889B1 (12) United States Patent (10) Patent No.: US 6,395,889 B1 Robison (45) Date of Patent: May 28, 2002 (54) NUCLEIC ACID MOLECULES ENCODING WO WO-98/56804 A1 * 12/1998 ........... CO7H/21/02 HUMAN PROTEASE HOMOLOGS WO WO-99/0785.0 A1 * 2/1999 ... C12N/15/12 WO WO-99/37660 A1 * 7/1999 ........... CO7H/21/04 (75) Inventor: fish E. Robison, Wilmington, MA OTHER PUBLICATIONS Vazquez, F., et al., 1999, “METH-1, a human ortholog of (73) Assignee: Millennium Pharmaceuticals, Inc., ADAMTS-1, and METH-2 are members of a new family of Cambridge, MA (US) proteins with angio-inhibitory activity', The Journal of c: - 0 Biological Chemistry, vol. 274, No. 33, pp. 23349–23357.* (*) Notice: Subject to any disclaimer, the term of this Descriptors of Protease Classes in Prosite and Pfam Data patent is extended or adjusted under 35 bases. U.S.C. 154(b) by 0 days. * cited by examiner (21) Appl. No.: 09/392, 184 Primary Examiner Ponnathapu Achutamurthy (22) Filed: Sep. 9, 1999 ASSistant Examiner William W. Moore (51) Int. Cl." C12N 15/57; C12N 15/12; (74) Attorney, Agent, or Firm-Alston & Bird LLP C12N 9/64; C12N 15/79 (57) ABSTRACT (52) U.S. Cl. .................... 536/23.2; 536/23.5; 435/69.1; 435/252.3; 435/320.1 The invention relates to polynucleotides encoding newly (58) Field of Search ............................... 536,232,235. identified protease homologs. The invention also relates to 435/6, 226, 69.1, 252.3 the proteases. The invention further relates to methods using s s s/ - - -us the protease polypeptides and polynucleotides as a target for (56) References Cited diagnosis and treatment in protease-mediated disorders. -

In Escherichia Coli (Synthetic Oligonucleotide/Gene Expression/Industrial Enzyme) J
Proc. Nati Acad. Sci. USA Vol. 80, pp. 3671-3675, June 1983 Biochemistry Synthesis of calf prochymosin (prorennin) in Escherichia coli (synthetic oligonucleotide/gene expression/industrial enzyme) J. S. EMTAGE*, S. ANGALt, M. T. DOELt, T. J. R. HARRISt, B. JENKINS*, G. LILLEYt, AND P. A. LOWEt Departments of *Molecular Genetics, tMolecular Biology, and tFermentation Development, Celltech Limited, 250 Bath Road, Slough SL1 4DY, Berkshire, United Kingdom Communicated by Sydney Brenner, March 23, 1983 ABSTRACT A gene for calf prochymosin (prorennin) has been maturation conditions and remained insoluble on neutraliza- reconstructed from chemically synthesized oligodeoxyribonucleo- tion, it was possible that purification as well as activation could tides and cloned DNA copies of preprochymosin mRNA. This gene be achieved. has been inserted into a bacterial expression plasmid containing We describe here the construction of E. coli plasmids de- the Escherichia coli tryptophan promoter and a bacterial ribo- signed to express the prochymosin gene from the trp promoter some binding site. Induction oftranscription from the tryptophan and the isolation and conversion of this prochymosin to en- promoter results in prochymosin synthesis at a level of up to 5% active of total protein. The enzyme has been purified from bacteria by zymatically chymosin. extraction with urea and chromatography on DEAE-celiulose and MATERIALS AND METHODS converted to enzymatically active chymosin by acidification and neutralization. Bacterially produced chymosin is as effective in Materials. DNase I, pepstatin A, and phenylmethylsulfonyl clotting milk as the natural enzyme isolated from calf stomach. fluoride were obtained from Sigma. Calf prochymosin (Mr 40,431) and chymosin (Mr 35,612) were purified from stomachs Chymosin (rennin) is an aspartyl proteinase found in the fourth from 1-day-old calves (1). -

Membrane Proteins • Cofactors – Plimstex • Membranes • Dna • Small Molecules/Gas • Large Complexes
Structural mass spectrometry hydrogen/deuterium exchange Petr Man Structural Biology and Cell Signalling Institute of Microbiology, Czech Academy of Sciences Structural biology methods Low-resolution methods High-resolution methods Rigid SAXS IR Raman CD ITC MST Cryo-EM AUC SPR MS X-ray crystallography Chemical cross-linking H/D exchange Native ESI + ion mobility Oxidative labelling Small Large NMR Dynamic Structural biology approaches Simple MS, quantitative MS Cross-linking, top-down, native MS+dissociation native MS+ion mobility Cross-linking Structural MS What can we get using mass spectrometry IM – ion mobility CXL – chemical cross-linking AP – afinity purification OFP – oxidative footprinting HDX – hydrogen/deuterium exchange ISOTOPE EXCHANGE IN PROTEINS 1H 2H 3H occurence [%] 99.988 0.0115 trace 5 …Kaj Ulrik Linderstrøm-Lang „Cartesian diver“ Proteins are migrating in tubes with density gradient until they stop at the point where the densities are equal 1H 2H 3H % 99.9885 0.0115 trace density [g/cm3] 1.000 1.106 1.215 Methods of detection IR: β-: NMR: 1 n = 1.6749 × 10-27 kg MS: 1H 2H 3H výskyt% [%] 99.9885 0.0115 trace hustotadensity vody [g/cm [g/cm3] 3] 1.000 1.106 1.215 jadernýspinspin ½+ 1+ ½+ mass [u] 1.00783 2.01410 3.01605 Factors affecting H/D exchange hydrogen bonding solvent accessibility Factors affecting H/D exchange Side chains (acidity, steric shielding) Bai et al.: Proteins (1993) Glasoe, Long: J. Phys. Chem. (1960) Factors affecting H/D exchange – side chain effects Inductive effect – electron density is Downward shift due to withdrawn from peptide steric hindrance effect of bond (S, O). -

(12) Patent Application Publication (10) Pub. No.: US 2016/0346364 A1 BRUNS Et Al
US 2016.0346364A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2016/0346364 A1 BRUNS et al. (43) Pub. Date: Dec. 1, 2016 (54) MEDICAMENT AND METHOD FOR (52) U.S. Cl. TREATING INNATE IMMUNE RESPONSE CPC ........... A61K 38/488 (2013.01); A61K 38/482 DISEASES (2013.01); C12Y 304/23019 (2013.01); C12Y 304/21026 (2013.01); C12Y 304/23018 (71) Applicant: DSM IPASSETS B.V., Heerlen (NL) (2013.01); A61K 9/0053 (2013.01); C12N 9/62 (2013.01); A23L 29/06 (2016.08); A2ID 8/042 (72) Inventors: Maaike Johanna BRUINS, Kaiseraugst (2013.01); A23L 5/25 (2016.08); A23V (CH); Luppo EDENS, Kaiseraugst 2002/00 (2013.01) (CH); Lenneke NAN, Kaiseraugst (CH) (57) ABSTRACT (21) Appl. No.: 15/101,630 This invention relates to a medicament or a dietary Supple (22) PCT Filed: Dec. 11, 2014 ment comprising the Aspergillus niger aspergilloglutamic peptidase that is capable of hydrolyzing plant food allergens, (86). PCT No.: PCT/EP2014/077355 and more particularly, alpha-amylase/trypsin inhibitors, thereby treating diseases due to an innate immune response S 371 (c)(1), in humans, and/or allowing to delay the onset of said (2) Date: Jun. 3, 2016 diseases. The present invention relates to the discovery that (30) Foreign Application Priority Data the Aspergillus niger aspergilloglutamic peptidase is capable of hydrolyzing alpha-amylase/trypsin inhibitors that are Dec. 11, 2013 (EP) .................................. 13196580.8 present in wheat and related cereals said inhibitors being strong inducers of innate immune response. Furthermore, Publication Classification the present invention relates to a method for hydrolyzing alpha-amylase/trypsin inhibitors comprising incubating a (51) Int. -

Cloning and in Vitro-Transcription of Chymosin Gene in E. Coli
The Open Nutraceuticals Journal, 2010, 3, 63-68 63 Open Access Cloning and In Vitro-Transcription of Chymosin Gene in E. coli S.A. El-Sohaimy1 Elsayed. E, Hafez2,* and M.A. El-Saadani3 1Food Science and Technology Department, Arid land Research Institute, Alexandria, Egypt 2Plant Molecular Pathology Department, Arid land Research Institute, Alexandria, Egypt 3Mubarak City for Scientific Research and Technology Applications, Alexandria, Egypt Abstract: Chymosin, commonly known as rennin, is the main milk-coagulating enzyme available in rennet. RNA was extracted from the abomasum of a suckling calf water buffalo and was subjected to RT-PCR using degenerate primers to amplify 850bp of the chymosin gene. The sequence was aligned with 19 different mammals' chymosin genes. The sequence revealed that there is a similarity to them ranging from 64% to 98%. The purified recombinant proteins were obtained from the transformed E. coli and yeast. The clotting activity of both of the resulting proteins was examined compared to the commercial peers. It was noticed that the concentration of the purified protein ranged from 15,000 to 40,000 MCU. Therefore, the activity of the obtained proteins was the same and it was 105% when compared to the commercial peer. Having examined the cytotoxicity of the purified proteins, the results revealed no toxicity. We can conclude that the obtained recombinant protein is more active and safe even when expressed in bacteria rather than yeast. Keywords: Chymosin, Milk Clotting, Recombinant E. coli and Recombinant Protein. INTRODUCTION Many microorganisms are known to produce rennet-like proteinases which can replace the calf rennet. -

Progress in the Field of Aspartic Proteinases in Cheese Manufacturing
Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering Sirma Yegin, Peter Dekker To cite this version: Sirma Yegin, Peter Dekker. Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering. Dairy Science & Technology, EDP sciences/Springer, 2013, 93 (6), pp.565-594. 10.1007/s13594-013-0137-2. hal-01201447 HAL Id: hal-01201447 https://hal.archives-ouvertes.fr/hal-01201447 Submitted on 17 Sep 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Dairy Sci. & Technol. (2013) 93:565–594 DOI 10.1007/s13594-013-0137-2 REVIEW PAPER Progress in the field of aspartic proteinases in cheese manufacturing: structures, functions, catalytic mechanism, inhibition, and engineering Sirma Yegin & Peter Dekker Received: 25 February 2013 /Revised: 16 May 2013 /Accepted: 21 May 2013 / Published online: 27 June 2013 # INRA and Springer-Verlag France 2013 Abstract Aspartic proteinases are an important class of proteinases which are widely used as milk-coagulating agents in industrial cheese production. They are available from a wide range of sources including mammals, plants, and microorganisms. -
![A Label-Free Cellular Proteomics Approach to Decipher the Antifungal Action of Dimiq, a Potent Indolo[2,3- B]Quinoline Agent, Against Candida Albicans Biofilms](https://docslib.b-cdn.net/cover/9827/a-label-free-cellular-proteomics-approach-to-decipher-the-antifungal-action-of-dimiq-a-potent-indolo-2-3-b-quinoline-agent-against-candida-albicans-biofilms-409827.webp)
A Label-Free Cellular Proteomics Approach to Decipher the Antifungal Action of Dimiq, a Potent Indolo[2,3- B]Quinoline Agent, Against Candida Albicans Biofilms
A Label-Free Cellular Proteomics Approach to Decipher the Antifungal Action of DiMIQ, a Potent Indolo[2,3- b]Quinoline Agent, against Candida albicans Biofilms Robert Zarnowski 1,2*, Anna Jaromin 3*, Agnieszka Zagórska 4, Eddie G. Dominguez 1,2, Katarzyna Sidoryk 5, Jerzy Gubernator 3 and David R. Andes 1,2 1 Department of Medicine, School of Medicine & Public Health, University of Wisconsin-Madison, Madison, WI 53706, USA; [email protected] (E.G.D.); [email protected] (D.R.A.) 2 Department of Medical Microbiology, School of Medicine & Public Health, University of Wisconsin-Madison, Madison, WI 53706, USA 3 Department of Lipids and Liposomes, Faculty of Biotechnology, University of Wroclaw, 50-383 Wroclaw, Poland; [email protected] 4 Department of Medicinal Chemistry, Jagiellonian University Medical College, 30-688 Cracow, Poland; [email protected] 5 Department of Pharmacy, Cosmetic Chemicals and Biotechnology, Team of Chemistry, Łukasiewicz Research Network-Industrial Chemistry Institute, 01-793 Warsaw, Poland; [email protected] * Correspondence: [email protected] (R.Z.); [email protected] (A.J.); Tel.: +1-608-265-8578 (R.Z.); +48-71-3756203 (A.J.) Label-Free Cellular Proteomics of Candida albicans biofilms treated with DiMIQ Identified Proteins Accession # Alternate ID Gene names (ORF ) WT DIMIQ Z SCORE Proteins induced by DiMIQ Arginase (EC 3.5.3.1) A0A1D8PP00 CAR1 CAALFM_C504490CA 0.000 6.648 drug induced Glucan 1,3-beta-glucosidase BGL2 (EC 3.2.1.58) (Exo-1Q5AMT2 BGL2 CAALFM_C402250CA -

Serine Proteases with Altered Sensitivity to Activity-Modulating
(19) & (11) EP 2 045 321 A2 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 08.04.2009 Bulletin 2009/15 C12N 9/00 (2006.01) C12N 15/00 (2006.01) C12Q 1/37 (2006.01) (21) Application number: 09150549.5 (22) Date of filing: 26.05.2006 (84) Designated Contracting States: • Haupts, Ulrich AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 51519 Odenthal (DE) HU IE IS IT LI LT LU LV MC NL PL PT RO SE SI • Coco, Wayne SK TR 50737 Köln (DE) •Tebbe, Jan (30) Priority: 27.05.2005 EP 05104543 50733 Köln (DE) • Votsmeier, Christian (62) Document number(s) of the earlier application(s) in 50259 Pulheim (DE) accordance with Art. 76 EPC: • Scheidig, Andreas 06763303.2 / 1 883 696 50823 Köln (DE) (71) Applicant: Direvo Biotech AG (74) Representative: von Kreisler Selting Werner 50829 Köln (DE) Patentanwälte P.O. Box 10 22 41 (72) Inventors: 50462 Köln (DE) • Koltermann, André 82057 Icking (DE) Remarks: • Kettling, Ulrich This application was filed on 14-01-2009 as a 81477 München (DE) divisional application to the application mentioned under INID code 62. (54) Serine proteases with altered sensitivity to activity-modulating substances (57) The present invention provides variants of ser- screening of the library in the presence of one or several ine proteases of the S1 class with altered sensitivity to activity-modulating substances, selection of variants with one or more activity-modulating substances. A method altered sensitivity to one or several activity-modulating for the generation of such proteases is disclosed, com- substances and isolation of those polynucleotide se- prising the provision of a protease library encoding poly- quences that encode for the selected variants. -

Crystal Structures of Native and Inhibitedforms of Human Cathepsin
Proc. Natl. Acad. Sci. USA Vol. 90, pp. 6796-6800, July 1993 Biochemustry Crystal structures of native and inhibited forms of human cathepsin D: Implications for lysosomal targeting and drug design (aspartic protcase/N-linked oligosaccharide/pepstatin A) ERic T. BALDWIN*, T. NARAYANA BHAT*, SERGEI GULNIK*, MADHUSOODAN V. HOSUR*t, RAYMOND C. SOWDER Il, RAUL E. CACHAU*, JACK COLLINS*, ABELARDO M. SILVA*, AND JOHN W. ERICKSON*§ *Structural Biochemistry Program, Frederick Biomedical Supercomputing Center and tAIDS Vaccine Program, Program Resources Inc./DynCorp, National Cancer Institute-Frederick Cancer Research and Development Center, Frederick, MD 21702 Communicated by David R. Davies, March 24, 1993 (receivedfor review February 4, 1993) ABSTRACT Cathepsin D (EC 3.4.23.5) is a lysosomal duced in the vicinity of the growing tumor, may degrade the protease suspected to play important roles in protein catabo- extracellular matrix and thereby promote the escape of lism, antigen processing, degenerative diseases, and breast cancer cells to the lymphatic and circulatory systems and cancer progresson. Determination of the crystal structures of enhance the invasion of new tissues (17, 18). The design of cathepsin D and a complex with pepstatin at 2.5 A resolution potent and specific inhibitors of cathepsin D will aid the provides insights into inhibitor binding and lysosomal targeting further elucidation of the roles of this enzyme in human for this two-chain, N-glycosylated aspartic protease. Compar- disease. We previously described the purification and crys- ison with the structures of a complex of pepstatin bound to tallization ofhuman cathepsin D from liver (3); similar studies rhizopuspepsin and with a human renin-bihbitor complex have been reported recently for cathepsin D isolated from revealed differences in subsite structures and inhibitor-enzyme bovine liver (19) and human spleen (20). -

Camel and Bovine Chymosin: the Relationship Crystallography Between Their Structures and Cheese-Making ISSN 0907-4449 Properties
research papers Acta Crystallographica Section D Biological Camel and bovine chymosin: the relationship Crystallography between their structures and cheese-making ISSN 0907-4449 properties Jesper Langholm Jensen,a,b Bovine and camel chymosin are aspartic peptidases that are Received 19 November 2012 Anne Mølgaard,a Jens-Christian used industrially in cheese production. They cleave the Accepted 31 January 2013 Navarro Poulsen,a Phe105-Met106 bond of the milk protein -casein, releasing b Marianne Kirsten Harboe, its predominantly negatively charged C-terminus, which leads PDB References: bovine Jens Bæk Simonsen,c‡ to the separation of the milk into curds and whey. Despite chymosin, 4aa8; camel chymosin, 4aa9 Andrea Maria Lorentzen,d Karin having 85% sequence identity, camel chymosin shows a 70% higher milk-clotting activity than bovine chymosin towards Hjernø,d Johannes M. van den bovine milk. The activities, structures, thermal stabilities and Brink,b Karsten Bruun Qvistb and a glycosylation patterns of bovine and camel chymosin obtained Sine Larsen * by fermentation in Aspergillus niger have been examined. Different variants of the enzymes were isolated by hydro- aDepartment of Chemistry, University of phobic interaction chromatography and showed variations Copenhagen, Denmark, bChr. Hansen A/S, in their glycosylation, N-terminal sequences and activities. Bøge Alle´ 10-12, DK-2970 Hørsholm, Glycosylation at Asn291 and the loss of the first three residues c Denmark, Nanobioscience, Department of of camel chymosin significantly decreased its activity. Thermal Basic Sciences and Environment, University of Copenhagen, Denmark, and dInstitute of differential scanning calorimetry revealed a slightly higher Biochemistry and Molecular Biology, thermal stability of camel chymosin compared with bovine University of Southern Denmark, Denmark chymosin. -

Penicillopepsin-JT2, a Recombinant Enzyme from Penicillium Janthinellum and the Contribution of a Hydrogen Bond in Subsite S3 to Kcat
Protein Science ~2000!, 9:991–1001. Cambridge University Press. Printed in the USA. Copyright © 2000 The Protein Society Penicillopepsin-JT2, a recombinant enzyme from Penicillium janthinellum and the contribution of a hydrogen bond in subsite S3 to kcat QING-NA CAO,1,3 MARLENE STUBBS,1 KENNY Q.P. NGO,1 MICHAEL WARD,2 ANNIE CUNNINGHAM,1 EMIL F. PAI,1 GUANG-CHOU TU,1,3 and THEO HOFMANN1 1 Department of Biochemistry, University of Toronto, Toronto, Ontario M5S 1A8, Canada 2 Genencor International, Inc., 925 Page Mill Road, Palo Alto, California 94304-1013 ~Received August 30, 1999; Final Revision February 7, 2000; Accepted March 10, 2000! Abstract The nucleotide sequence of the gene ~ pepA! of a zymogen of an aspartic proteinase from Penicillium janthinellum with a 71% identity in the deduced amino acid sequence to penicillopepsin ~which we propose to call penicillopepsin-JT1! has been determined. The gene consists of 60 codons for a putative leader sequence of 20 amino acid residues, a sequence of about 150 nucleotides that probably codes for an activation peptide and a sequence with two introns that codes for the active aspartic proteinase. This gene, inserted into the expression vector pGPT-pyrG1, was expressed in an aspartic proteinase-free strain of Aspergillus niger var. awamori in high yield as a glycosylated form of the active enzyme that we call penicillopepsin-JT2. After removal of the carbohydrate component with endoglycosidase H, its relative molecular mass is between 33,700 and 34,000. Its kinetic properties, especially the rate-enhancing effects of the presence of alanine residues in positions P3 and P29 of substrates, are similar to those of penicillopepsin-JT1, endothia- pepsin, rhizopuspepsin, and pig pepsin.