Molecular Detection of Bacteria from a Possible Material Oral Origin in Neonatal Gastric Aspirates Obtained from Complicated Pregnancies Gonzales Marin, Cecilia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bartonella Apis Sp. Nov., a Honey Bee Gut Symbiont of the Class Alphaproteobacteria
Serveur Academique´ Lausannois SERVAL serval.unil.ch Author Manuscript Faculty of Biology and Medicine Publication This paper has been peer-reviewed but does not include the final publisher proof-corrections or journal pagination. Published in final edited form as: Title: Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Authors: Keˇsnerov´aL, Moritz R, Engel P Journal: International journal of systematic and evolutionary microbiology Year: 2016 Jan Issue: 66 Volume: 1 Pages: 414-21 DOI: 10.1099/ijsem.0.000736 In the absence of a copyright statement, users should assume that standard copyright protection applies, unless the article contains an explicit statement to the contrary. In case of doubt, contact the journal publisher to verify the copyright status of an article. 1 Bartonella apis sp. nov., a honey bee gut symbiont of the 2 class Alphaproteobacteria 3 4 Lucie Kešnerová, Roxane Moritz, Philipp Engel* 5 6 Department of Fundamental Microbiology, University of Lausanne, CH-1015 7 Lausanne, Switzerland 8 9 Running title: Description of a bee gut symbiont 10 11 *Correspondence: 12 Prof. Philipp Engel 13 Department of Fundamental Microbiology 14 University of Lausanne, CH-1015 Lausanne, Switzerland 15 Tel.: +41 (0)21 692 56 12 16 e-mail: [email protected] 17 18 Category: New Taxa – Proteobacteria 19 Keywords: Apis mellifera; insect; Bartonella; gut microbiota; Alpha-1 20 21 Sequence deposition: The 16S rRNA gene sequences and protein-coding gene 22 sequences of the bacterial strains PEB0122T, PEB0149, PEB0150, BBC0104, and 23 BBC0108 from Apis mellifera, and the uncultured Rhizobiales bacterium from 24 Herpagnathos saltator are deposited in GenBank with accession numbers KP987849 25 – KP987886 and KT315729 – KT315734. -
Additional File 1
Additional file 1 Microbial succession during the transition from active to inactive stages of deep-sea hydrothermal vent sulfide chimneys Jialin Hou, Stefan M. Sievert, Yinzhao Wang, Jeff S. Seewald, Vengadesh Perumal Natarajan Fengping Wang*, Xiang Xiao* a b 100 100 75 75 Taxa Alphaproteobacteria Taxa Betaproteobacteria Aquificae Deinococcus-Thermus Campylobacteria Deltaproteobacteria 50 Euryarchaeata 50 Euryarchaeata Others Gammaproteobacteria Percentage Percentage Thermodesulfobacterium Muproteobacteria Unclassified Nitrospirae Others Unclassified 25 25 0 0 aclA aclB cdhA cdhC napA napB norB nosZ sat soxA soxC soxY soxZ sqr aprA aprB dsrA dsrB narG nirB PRK rbcL rbcS Gene Gene Figure S1 Taxonomic classification of key functional genes retrieved from the L- and M-vent chimeny.(a) The key genes enriched in the active L-vent chimney. (b) The key genes enriched in the recently inactive M-vent chimney. Reductive bacterial type Reductive archaeal type Chlorobi group Archaeoglobus(3) Oxidative bacterial type Crenarchaeota Alphaproteobacteria(1/16) Nitrospirae(12) Acidobacteria(1/14) Betaproteobacteria Firmicutes groups Gammaproteobacteria (20) Tree scale: 1 Deltaproteobacteria(9/10) Reductive bacterial type Figure S2 Maximum-likelihood phylogeny of dsrA genes retrieved from L- and M-vent chimney. The red branches represent the dsrA genes recovered from active the L-vent chimney, while the blue ones are those from recently inactive M-vent chimney. Numbers of dsrA gene for each sample are displayed in the parenthesis after the clade name. -

View Tickborne Diseases Sample Report
1360 Bayport Ave, Ste B. San Carlos, CA 94070 1(866) 364-0963 | [email protected] | www. vibrant-wellness.com PATIENT PROVIDER NAME: DEMO REPORT GENDER: Male PRACTICE NAME: Vibrant IT4 Practice DATE OF BIRTH: 04/14/1998 AGE: 22 PROVIDER NAME: Demo Client, DDD (999994) ADDRESS: TEST STREET, TEST CITY, KY- 42437. ACCESSION ID: 2009220006 PHLEBOTOMIST: 607 SPECIMEN COLLECTION TIME: 09-21-2020 11:14 SPECIMEN RECEIVED TIME: 09-22-2020 05:14 FINAL REPORT TIME: 09-25-2020 15:56 FASTING: FASTING Your Vibrant Wellness TickBorne 2.0 panel results are enclosed. These results are intended to aid in the diagnosis of tickborne diseases by your healthcare provider. The Vibrant Tickborne Diseases panel tests for IgG and IgM antibodies for Borreliosis/Lyme disease as well as co-infection(s) and opportunistic infections with other tick-borne illnesses along with detection of DNA of the species causing these infections. The Vibrant Immunochip test is a semiquantitative assay that detects IgG and IgM antibodies in human serum. The PCR Test is a real-time PCR Assay designed for qualitative detection of infectious group- specific DNA in clinical samples. Interpretation of Report: The test results of antibody levels to the individual antigens are calculated by comparing the average intensity of the individual antibody to that of a reference population and cut-off chosen for each protein. Reference ranges have been established using a well characterized set of more than 300 serum samples and antibodies to specific bacteria tested. The results are displayed as In Control, Moderate, or High Risk.for each antigen tested. -

The Mysterious Orphans of Mycoplasmataceae
The mysterious orphans of Mycoplasmataceae Tatiana V. Tatarinova1,2*, Inna Lysnyansky3, Yuri V. Nikolsky4,5,6, and Alexander Bolshoy7* 1 Children’s Hospital Los Angeles, Keck School of Medicine, University of Southern California, Los Angeles, 90027, California, USA 2 Spatial Science Institute, University of Southern California, Los Angeles, 90089, California, USA 3 Mycoplasma Unit, Division of Avian and Aquatic Diseases, Kimron Veterinary Institute, POB 12, Beit Dagan, 50250, Israel 4 School of Systems Biology, George Mason University, 10900 University Blvd, MSN 5B3, Manassas, VA 20110, USA 5 Biomedical Cluster, Skolkovo Foundation, 4 Lugovaya str., Skolkovo Innovation Centre, Mozhajskij region, Moscow, 143026, Russian Federation 6 Vavilov Institute of General Genetics, Moscow, Russian Federation 7 Department of Evolutionary and Environmental Biology and Institute of Evolution, University of Haifa, Israel 1,2 [email protected] 3 [email protected] 4-6 [email protected] 7 [email protected] 1 Abstract Background: The length of a protein sequence is largely determined by its function, i.e. each functional group is associated with an optimal size. However, comparative genomics revealed that proteins’ length may be affected by additional factors. In 2002 it was shown that in bacterium Escherichia coli and the archaeon Archaeoglobus fulgidus, protein sequences with no homologs are, on average, shorter than those with homologs [1]. Most experts now agree that the length distributions are distinctly different between protein sequences with and without homologs in bacterial and archaeal genomes. In this study, we examine this postulate by a comprehensive analysis of all annotated prokaryotic genomes and focusing on certain exceptions. -

A New Symbiotic Lineage Related to Neisseria and Snodgrassella Arises from the Dynamic and Diverse Microbiomes in Sucking Lice
bioRxiv preprint doi: https://doi.org/10.1101/867275; this version posted December 6, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. A new symbiotic lineage related to Neisseria and Snodgrassella arises from the dynamic and diverse microbiomes in sucking lice Jana Říhová1, Giampiero Batani1, Sonia M. Rodríguez-Ruano1, Jana Martinů1,2, Eva Nováková1,2 and Václav Hypša1,2 1 Department of Parasitology, Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic 2 Institute of Parasitology, Biology Centre, ASCR, v.v.i., České Budějovice, Czech Republic Author for correspondence: Václav Hypša, Department of Parasitology, University of South Bohemia, České Budějovice, Czech Republic, +42 387 776 276, [email protected] Abstract Phylogenetic diversity of symbiotic bacteria in sucking lice suggests that lice have experienced a complex history of symbiont acquisition, loss, and replacement during their evolution. By combining metagenomics and amplicon screening across several populations of two louse genera (Polyplax and Hoplopleura) we describe a novel louse symbiont lineage related to Neisseria and Snodgrassella, and show its' independent origin within dynamic lice microbiomes. While the genomes of these symbionts are highly similar in both lice genera, their respective distributions and status within lice microbiomes indicate that they have different functions and history. In Hoplopleura acanthopus, the Neisseria-related bacterium is a dominant obligate symbiont universally present across several host’s populations, and seems to be replacing a presumably older and more degenerated obligate symbiont. -

Tick-Borne Pathogens in Removed Ticks Veneto, Northeastern Italy
Tick-borne pathogens in removed ticks Veneto, northeastern Italy: A cross-sectional investigation Anna Beltrame, Maureen Laroche, Monica Degani, Francesca Perandin, Zeno Bisoffi, Didier Raoult, Philippe Parola To cite this version: Anna Beltrame, Maureen Laroche, Monica Degani, Francesca Perandin, Zeno Bisoffi, et al.. Tick- borne pathogens in removed ticks Veneto, northeastern Italy: A cross-sectional investigation. Travel Medicine and Infectious Disease, Elsevier, 2018, 26, pp.58-61. 10.1016/j.tmaid.2018.08.008. hal- 01970220 HAL Id: hal-01970220 https://hal.archives-ouvertes.fr/hal-01970220 Submitted on 10 Apr 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Travel Medicine and Infectious Disease 26 (2018) 58–61 Contents lists available at ScienceDirect Travel Medicine and Infectious Disease journal homepage: www.elsevier.com/locate/tmaid Tick-borne pathogens in removed ticks Veneto, northeastern Italy: A cross- sectional investigation T ∗ Anna Beltramea, , Maureen Larocheb, Monica Degania, Francesca Perandina, Zeno Bisoffia, Didier Raoultc, Philippe Parolab a Centre for Tropical Diseases, IRCCS Sacro Cuore Don Calabria Hospital, Via Sempreboni 5, 37024, Negrar, Italy b Aix Marseille Univ, AP-HM, SSA, VITROME, IHU-Méditerranée Infection, 19-21 Bd Jean Moulin, 13005, Marseille, France c Aix Marseille Univ, AP-HM, MEPHI, IHU-Méditerranée Infection, 19-21 Bd Jean Moulin, 13005, Marseille, France ARTICLE INFO ABSTRACT Keywords: Background: In Italy, the incidence of tick-borne diseases in humans is underestimated, as they are not ob- Tick-borne diseases ligatorily notifiable. -

Correlation Between the Oral Microbiome and Brain Resting State Connectivity in Smokers
bioRxiv preprint doi: https://doi.org/10.1101/444612; this version posted October 16, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Correlation between the oral microbiome and brain resting state connectivity in smokers Dongdong Lin1, Kent Hutchison2, Salvador Portillo3, Victor Vegara1, Jarod Ellingson2, Jingyu Liu1,3, Amanda Carroll-Portillo3,* ,Vince D. Calhoun1,3,* 1The Mind Research Network, Albuquerque, New Mexico, 87106 2University of Colorado Boulder, Boulder, CO 3University of New Mexico, Department of Electrical and Computer Engineering, Albuquerque, New Mexico, 87106 * authors contributed equally to the work. Abstract Recent studies have shown a critical role for the gastrointestinal microbiome in brain and behavior via a complex gut–microbiome–brain axis, however, the influence of the oral microbiome in neurological processes is much less studied, especially in response to the stimuli in the oral microenvironment such as smoking. Additionally, given the complex structural and functional networks in brain system, our knowledge about the relationship between microbiome and brain functions on specific brain circuits is still very limited. In this pilot work, we leverage next generation microbial sequencing with functional MRI techniques to enable the delineation of microbiome-brain network links as well as their relations to cigarette smoking. Thirty smokers and 30 age- and sex- matched non-smokers were recruited for measuring both microbial community and brain functional networks. Statistical analyses were performed to demonstrate the influence of smoking on: the taxonomy and abundance of the constituents within the oral microbial community, brain functional network connectivity, and associations between microbial shifts and the brain signaling network. -

Co-Infection of Bacteria and Protozoan Parasites in Ixodes Ricinus Nymphs Collected in the Alsace Region, France
Ticks and Tick-borne Diseases 10 (2019) 101241 Contents lists available at ScienceDirect Ticks and Tick-borne Diseases journal homepage: www.elsevier.com/locate/ttbdis Short communication Co-infection of bacteria and protozoan parasites in Ixodes ricinus nymphs T collected in the Alsace region, France Amira Nebbaka,b, Handi Dahmanaa, Lionel Almerasa,c, Didier Raoultd, Nathalie Boulangere,f, ⁎ Benoit Jaulhace,f, Oleg Mediannikovd, Philippe Parolaa,d, a Aix Marseille Univ, IRD, AP-HM, SSA, VITROME, Marseille, France b Centre de Recherche Scientifique et Technique en Analyses Physico-Chimiques (CRAPC). Zone Industrielle, BP 384 Bou-Ismail, Tipaza Algeria c Unité de Parasitologie et Entomologie, Département des Maladies Infectieuses, Institut de Recherche Biomédicale des Armées, Marseille, France d IHU-Méditerranée Infection, Marseille, France e Centre National de Reference Borrelia, Centre Hospitalier Universitaire, Strasbourg, France f EA7290Virulence bactérienne précoce: groupe borréliose de Lyme, Facultés de pharmacie et de médecine, Université de Strasbourg, France ARTICLE INFO ABSTRACT Keywords: Fifty nymphal Ixodes ricinus ticks collected in Alsace, France, identified by morphological criteria and using Ixodes ricinus MALDI-TOF MS, were tested by PCR to detect tick-associated bacteria and protozoan parasites. Seventy percent Co-infection (35/50) of ticks contained at least one microorganism; 26% (9/35) contained two or more species. Several Bacterial pathogens human pathogens were identified including Borrelia burgdorferi s.s. (4%), Borrelia afzelii (2%), Borrelia garinii Protozoan parasites (2%), Borrelia valaisiana (4%), Borrelia miyamotoi (2%), Rickettsia helvetica (6%) and “Babesia venatorum” (2%). MALDI-TOF MS Bartonella spp. (10%) and a Wolbachia spp. (8%) were also detected. The most common co-infections involved Anaplasmataceae with Borrelia spp. -

Integrating Taxonomic, Functional, and Strain-Level Profiling of Diverse
TOOLS AND RESOURCES Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3 Francesco Beghini1†, Lauren J McIver2†, Aitor Blanco-Mı´guez1, Leonard Dubois1, Francesco Asnicar1, Sagun Maharjan2,3, Ana Mailyan2,3, Paolo Manghi1, Matthias Scholz4, Andrew Maltez Thomas1, Mireia Valles-Colomer1, George Weingart2,3, Yancong Zhang2,3, Moreno Zolfo1, Curtis Huttenhower2,3*, Eric A Franzosa2,3*, Nicola Segata1,5* 1Department CIBIO, University of Trento, Trento, Italy; 2Harvard T.H. Chan School of Public Health, Boston, United States; 3The Broad Institute of MIT and Harvard, Cambridge, United States; 4Department of Food Quality and Nutrition, Research and Innovation Center, Edmund Mach Foundation, San Michele all’Adige, Italy; 5IEO, European Institute of Oncology IRCCS, Milan, Italy Abstract Culture-independent analyses of microbial communities have progressed dramatically in the last decade, particularly due to advances in methods for biological profiling via shotgun metagenomics. Opportunities for improvement continue to accelerate, with greater access to multi-omics, microbial reference genomes, and strain-level diversity. To leverage these, we present bioBakery 3, a set of integrated, improved methods for taxonomic, strain-level, functional, and *For correspondence: phylogenetic profiling of metagenomes newly developed to build on the largest set of reference [email protected] (CH); sequences now available. Compared to current alternatives, MetaPhlAn 3 increases the accuracy of [email protected] (EAF); taxonomic profiling, and HUMAnN 3 improves that of functional potential and activity. These [email protected] (NS) methods detected novel disease-microbiome links in applications to CRC (1262 metagenomes) and †These authors contributed IBD (1635 metagenomes and 817 metatranscriptomes). -

Bacterial Diversity and Functional Analysis of Severe Early Childhood
www.nature.com/scientificreports OPEN Bacterial diversity and functional analysis of severe early childhood caries and recurrence in India Balakrishnan Kalpana1,3, Puniethaa Prabhu3, Ashaq Hussain Bhat3, Arunsaikiran Senthilkumar3, Raj Pranap Arun1, Sharath Asokan4, Sachin S. Gunthe2 & Rama S. Verma1,5* Dental caries is the most prevalent oral disease afecting nearly 70% of children in India and elsewhere. Micro-ecological niche based acidifcation due to dysbiosis in oral microbiome are crucial for caries onset and progression. Here we report the tooth bacteriome diversity compared in Indian children with caries free (CF), severe early childhood caries (SC) and recurrent caries (RC). High quality V3–V4 amplicon sequencing revealed that SC exhibited high bacterial diversity with unique combination and interrelationship. Gracillibacteria_GN02 and TM7 were unique in CF and SC respectively, while Bacteroidetes, Fusobacteria were signifcantly high in RC. Interestingly, we found Streptococcus oralis subsp. tigurinus clade 071 in all groups with signifcant abundance in SC and RC. Positive correlation between low and high abundant bacteria as well as with TCS, PTS and ABC transporters were seen from co-occurrence network analysis. This could lead to persistence of SC niche resulting in RC. Comparative in vitro assessment of bioflm formation showed that the standard culture of S. oralis and its phylogenetically similar clinical isolates showed profound bioflm formation and augmented the growth and enhanced bioflm formation in S. mutans in both dual and multispecies cultures. Interaction among more than 700 species of microbiota under diferent micro-ecological niches of the human oral cavity1,2 acts as a primary defense against various pathogens. Tis has been observed to play a signifcant role in child’s oral and general health. -

Caldimonas Taiwanensis Sp. Nov., a Amylase Producing Bacterium
ARTICLE IN PRESS Systematic and Applied Microbiology 28 (2005) 415–420 www.elsevier.de/syapm Caldimonas taiwanensis sp. nov., a amylase producing bacterium isolated from a hot spring Wen-Ming Chena,Ã, Jo-Shu Changb, Ching-Hsiang Chiua, Shu-Chen Changc, Wen-Chieh Chena, Chii-Ming Jianga aDepartment of Seafood Science, National Kaohsiung Marine University, No. 142, Hai-Chuan Rd. Nan-Tzu, Kaohsiung City 811, Taiwan bDepartment of Chemical Engineering, National Cheng Kung University, Tainan, Taiwan cTajen Institute of Technology, Yen-Pu, Pingtung, Taiwan Received 22 February 2005 Abstract During screening for amylase-producing bacteria, a strain designated On1T was isolated from a hot spring located in Pingtung area, which is near the very southern part of Taiwan. Cells of this organism were Gram-negative rods motile by means of a single polar flagellum. Optimum conditions for growthwere 55 1C and pH 7. Strain On1T grew well in minimal medium containing starchas thesole carbon source, and its extracellular products expressed amylase activity. The 16S rRNA gene sequence analysis indicates that strain On1T is a member of b-Proteobacteria. On the basis of a phylogenetic analysis of 16S rDNA sequences, DNA–DNA similarity data, physiological and biochemical characteristics, as well as fatty acid compositions, the organism belonged to the genus Caldimonas and represented a novel species within this genus. The predominant cellular fatty acids of strain On1T were 16:0 (about 30%), 18:1 o7c (about 20%) and summed feature 3 (16:1o7c or 15:0 iso 2OH or both[about 31%]). Its DNA base ratio was 65.9 mol% G+C. -
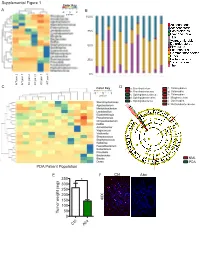
Mental Figure 1 Color Key a -2 0 2 B Z-Score 100%
Supplemental Figure 1 Color Key A -2 0 2 B z-score 100% 75% 50% 25% 0% KC pan 1 WT pan 3 WT KC pan 3 WT pan 2 WT pan 1 WT KC pan 2 C Color Key D a: Brevibacterium f: Chlamydiales b: Brevibacteriaceae g: Chlamydiia -3 0 3 z-score c: Sphingobacteriaceae h: Chlamydiae d: Sphingobacteriales i: Mogibacterium e: Sphingobacteriia j: Oscillospira k: Methylobacteriaceae NML Control Microb.-entrained MΦ PDA PDA Patient Population Control Microb.-entrained MΦ + Myd88i E F Ctrl Abx 350 * 300 250 200 150 40X 100 Tumor weight (mg) 50 0 x Ctrl Ab Supplemental Figure 2 A KC WT B ** * Actinobacteria * ** Bacteroidetes Cyanobacteria Deferribacteres * Firmicutes Proteobacteria % Relative abundance TM7 Others Time(wks) 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 Alpha Diversity Measure C E 60 KC WT 40 20 B. pseudolongum B. animalis 60 5 KC WT 0 B. adolescentis 40% Rel. abundance 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 20 Age (weeks) B. pseudolongum B. animalis 5 0 B. adolescentis % Rel. abundance 3 9 13 16 20 24 28 32 36 3 9 13 16 20 24 28 32 36 F Age (weeks) Week 3 Week 9 Week 13 p=0.678 p=0.02 p=0.385 Time(wks) 3 9 24 20 13 16 D 28 32 36 Week 13 KC WT Firmicutes; Ruminococcus Firmicutes; Dehalobacterium Alpha Diversity Measure Firmicutes; Oscillospira Bacteroidates; Odoribacter Axis.2 [12.7%] Actinobacteria; Bifidobacterium Axis.2 [23.8%] Axis.2 [24.7%] Week 16 Bacteroidetes; Bacteroidales Axis.1 [80.8%] Axis.1 [65.4%] Axis.1 [49.6%] Actinobacteria; Bifidobacterium Week 16 Week 20 Week 24 Week 20 p=0.339 p=0.036 p=0.021 Firmicutes; Dehalobacterium