(Siluriformes: Trichomycterus) at a Small Geographical Scale
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
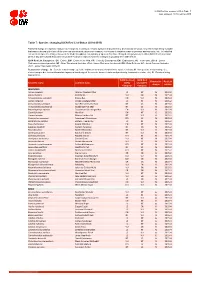
Table 7: Species Changing IUCN Red List Status (2014-2015)
IUCN Red List version 2015.4: Table 7 Last Updated: 19 November 2015 Table 7: Species changing IUCN Red List Status (2014-2015) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2014 (IUCN Red List version 2014.3) and 2015 (IUCN Red List version 2015-4) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered, EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2014) List (2015) change version Category Category MAMMALS Aonyx capensis African Clawless Otter LC NT N 2015-2 Ailurus fulgens Red Panda VU EN N 2015-4 -

Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi
Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 (http://www.accessscience.com/) Article by: Boschung, Herbert Department of Biological Sciences, University of Alabama, Tuscaloosa, Alabama. Gardiner, Brian Linnean Society of London, Burlington House, Piccadilly, London, United Kingdom. Publication year: 2014 DOI: http://dx.doi.org/10.1036/1097-8542.680400 (http://dx.doi.org/10.1036/1097-8542.680400) Content Morphology Euteleostei Bibliography Phylogeny Classification Additional Readings Clupeomorpha and Ostariophysi The most recent group of actinopterygians (rayfin fishes), first appearing in the Upper Triassic (Fig. 1). About 26,840 species are contained within the Teleostei, accounting for more than half of all living vertebrates and over 96% of all living fishes. Teleosts comprise 517 families, of which 69 are extinct, leaving 448 extant families; of these, about 43% have no fossil record. See also: Actinopterygii (/content/actinopterygii/009100); Osteichthyes (/content/osteichthyes/478500) Fig. 1 Cladogram showing the relationships of the extant teleosts with the other extant actinopterygians. (J. S. Nelson, Fishes of the World, 4th ed., Wiley, New York, 2006) 1 of 9 10/7/2015 1:07 PM Teleostei - AccessScience from McGraw-Hill Education http://www.accessscience.com/content/teleostei/680400 Morphology Much of the evidence for teleost monophyly (evolving from a common ancestral form) and relationships comes from the caudal skeleton and concomitant acquisition of a homocercal tail (upper and lower lobes of the caudal fin are symmetrical). This type of tail primitively results from an ontogenetic fusion of centra (bodies of vertebrae) and the possession of paired bracing bones located bilaterally along the dorsal region of the caudal skeleton, derived ontogenetically from the neural arches (uroneurals) of the ural (tail) centra. -

Multilocus Analysis of the Catfish Family Trichomycteridae (Teleostei: Ostario- Physi: Siluriformes) Supporting a Monophyletic Trichomycterinae
Accepted Manuscript Multilocus analysis of the catfish family Trichomycteridae (Teleostei: Ostario- physi: Siluriformes) supporting a monophyletic Trichomycterinae Luz E. Ochoa, Fabio F. Roxo, Carlos DoNascimiento, Mark H. Sabaj, Aléssio Datovo, Michael Alfaro, Claudio Oliveira PII: S1055-7903(17)30306-8 DOI: http://dx.doi.org/10.1016/j.ympev.2017.07.007 Reference: YMPEV 5870 To appear in: Molecular Phylogenetics and Evolution Received Date: 28 April 2017 Revised Date: 4 July 2017 Accepted Date: 7 July 2017 Please cite this article as: Ochoa, L.E., Roxo, F.F., DoNascimiento, C., Sabaj, M.H., Datovo, A., Alfaro, M., Oliveira, C., Multilocus analysis of the catfish family Trichomycteridae (Teleostei: Ostariophysi: Siluriformes) supporting a monophyletic Trichomycterinae, Molecular Phylogenetics and Evolution (2017), doi: http://dx.doi.org/10.1016/ j.ympev.2017.07.007 This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain. Multilocus analysis of the catfish family Trichomycteridae (Teleostei: Ostariophysi: Siluriformes) supporting a monophyletic Trichomycterinae Luz E. Ochoaa, Fabio F. Roxoa, Carlos DoNascimientob, Mark H. Sabajc, Aléssio -

Acanthopterygii, Bone, Eurypterygii, Osteology, Percomprpha
Research in Zoology 2014, 4(2): 29-42 DOI: 10.5923/j.zoology.20140402.01 Comparative Osteology of the Jaws in Representatives of the Eurypterygian Fishes Yazdan Keivany Department of Natural Resources (Fisheries Division), Isfahan University of Technology, Isfahan, 84156-83111, Iran Abstract The osteology of the jaws in representatives of 49 genera in 40 families of eurypterygian fishes, including: Aulopiformes, Myctophiformes, Lampridiformes, Polymixiiformes, Percopsiformes, Mugiliformes, Atheriniformes, Beloniformes, Cyprinodontiformes, Stephanoberyciformes, Beryciformes, Zeiformes, Gasterosteiformes, Synbranchiformes, Scorpaeniformes (including Dactylopteridae), and Perciformes (including Elassomatidae) were studied. Generally, in this group, the upper jaw consists of the premaxilla, maxilla, and supramaxilla. The lower jaw consists of the dentary, anguloarticular, retroarticular, and sesamoid articular. In higher taxa, the premaxilla bears ascending, articular, and postmaxillary processes. The maxilla usually bears a ventral and a dorsal articular process. The supramaxilla is present only in some taxa. The dentary is usually toothed and bears coronoid and posteroventral processes. The retroarticular is small and located at the posteroventral corner of the anguloarticular. Keywords Acanthopterygii, Bone, Eurypterygii, Osteology, Percomprpha following method for clearing and staining bone and 1. Introduction cartilage provided in reference [18]. A camera lucida attached to a Wild M5 dissecting stereomicroscope was used Despite the introduction of modern techniques such as to prepare the drawings. The bones in the first figure of each DNA sequencing and barcoding, osteology, due to its anatomical section are arbitrarily shaded and labeled and in reliability, still plays an important role in the systematic the others are shaded in a consistent manner (dark, medium, study of fishes and comprises a major percent of today’s and clear) to facilitate comparison among the taxa. -

Fuzzy Logic-Based Expert System for Native Fish Habitat Assessment in a Scarcity Information Context
Ecohydrology of Surface and Groundwater Dependent Systems: Concepts, Methods and Recent Developments 77 (Proc. of JS.1 at the Joint IAHS & IAH Convention, Hyderabad, India, September 2009). IAHS Publ. 328, 2009. Fuzzy logic-based expert system for native fish habitat assessment in a scarcity information context R. I. MEZA & H. X. VARGAS Civil Engineering Department, Water Resources and Environmental Division, University of Chile, Av. Blanco Encalada #2002, 3º piso, Santiago, Chile [email protected] Abstract Expert systems are typically developed to solve unclear application fields or problems. Such is the case for Chilean river ecosystems, and their fish fauna. In Chile, fish fauna studies are led almost exclusively by biologists, but no or little information is available about reproductive season, fertility, reproductive strategies, age, locomotive capacity, migration, trophic dynamic or niche, to name a few. Quantitative studies are required to analyse the impact of exotic over native species and the antropic effect over a population’s decrease. This information is essential to take appropriate conservation measures for each species and aquatic system. On the other hand, the lack of an adequate hydrometric network forces the use of complementary tools that introduce uncertainties, which are expensive and of difficult quantification. Due to the absence of gauged data for the study area, an expert system application methodology was coupled to the hydrological and hydraulic models, GR4J and HEC-RAS, respectively. Fish species information was taken from biological studies performed for the basin of the Bio Bio River where 17 fish species (13 native and 4 exotic, were found); furthermore, 7 species are classified as vulnerable and 7 are endangered. -

Biodiversidad Final.Pmd
Gayana 70(1): 100-113, 2006 ISSN 0717-652X ESTADO DE CONOCIMIENTO DE LOS PECES DULCEACUICOLAS DE CHILE CURRENT STATE OF KNOWLEDGE OF FRESHWATER FISHES OF CHILE Evelyn Habit1, Brian Dyer2 & Irma Vila3 1Unidad de Sistemas Acuáticos, Centro de Ciencias Ambientales EULA-Chile, Universidad de Concepción, Casilla 160-C, Concepción, Chile. [email protected] 2Escuela de Recursos Naturales, Universidad del Mar, Amunátegui 1838, Recreo, Viña del Mar, Chile. [email protected] 3Laboratorio de Limnología, Depto. Ciencias Ecológicas, Facultad de Ciencias, Universidad de Chile, Santiago, Chile. [email protected] RESUMEN La ictiofauna nativa de los sistemas límnicos de Chile se compone de 11 familias, 17 géneros y alrededor de 44 especies, incluyendo dos lampreas. De éstas, 81% son endémicas de la provincia biogeográfica chilena y 40% se encuentran clasificadas en peligro de extinción. Los grupos más representados corresponden a los órdenes Siluriformes (11 especies), Osmeriformes (9 especies) y Atheriniformes (7 especies). También están representados en Chile los ciclóstomos Petromyzontiformes (2 especies), y los teleósteos Characiformes (4 especies), Cyprinodontiformes (6 especies), Perciformes (4 especies) y Mugilifromes (1). Latitudinalmente, la mayor riqueza de especies ocurre en la zona centro-sur de la provincia Chilena, en tanto que los extremos norte y sur son de baja riqueza específica. Dado su origen, porcentaje de endemismo y retención de caracteres primitivos, este conjunto ictiofaunístico es de alto valor biogeográfico y de conservación. Existen sin embargo importantes vacíos de conocimiento sobre su sistemática, distribución y biología. PALABRAS CLAVES: Peces, sistemas dulceacuícolas, Chile. ABSTRACT The Chilean native freshwater ichthyofauna is composed of 11 families, 17 genera and about 44 species, including two lampreys. -

A New Species of Trichogenesfrom the Rio Itapemirim Drainage
Neotropical Ichthyology, 8(4):707-717, 2010 Copyright © 2010 Sociedade Brasileira de Ictiologia A new species of Trichogenes from the rio Itapemirim drainage, southeastern Brazil, with comments on the monophyly of the genus (Siluriformes: Trichomycteridae) Mário C. C. de Pinna1, José Luiz Helmer2, Heraldo A. Britski1 and Leandro Rodrigues Nunes2 A new species of the formerly monotypic genus Trichogenes is described from a high-altitude stream of the rio Itapemirim system, an isolated Atlantic drainage in the State of Espírito Santo, southeastern Brazil. Trichogenes claviger, new species, differs from all other trichomycterids by the sexually dimorphic posterior process of the opercle, much elongated in males; the terminal mouth; the deeply bifurcated anterior neural spines and the presence of a large anterodorsal claw-like process on the neural arches of the anterior four free vertebrae. The new species also differs from its only congener, T. longipinnis, by a number of additional traits, including the the lack of branched anal-fin rays in specimens of any size; the broader than long posterior nostril; the deeper head (head depth 72.9-86.6% HL); the presence of a fine dark line along the base of the anal fin; the lack of dark spots on cheeks; the shape of the interopercle; the presence of odontodes on a bony expansion on the posterodorsal margin of the interopercle; the fewer vertebrae (35); the absence of an antorbital; and the fewer pleural ribs (eight). Small juveniles of the new species are also strikingly different from those of all other Trichomycteridae, including T. longipinnis, having a very large lateral eye, an upturned mouth, and compressed head. -

Monograph of the Cyprinid Fis~Hes of the Genus Garra Hamilton (173)
MONOGRAPH OF THE CYPRINID FIS~HES OF THE GENUS GARRA HAMILTON By A. G. K. MENON, Zoologist, ,Zoological Surt1ey of India, Oalcutta. (With 1 Table, 29 Text-figs. and 6 Plates) CONTENTS Page I-Introduction 175 II-Purpose and general results 176 III-Methods and approaches 176 (a) The definition of Measurements 176 (b) The analysis of Intergradation 178 (c) The recognition of subspecies. 179 (d) Procedures in the paper 180 (e) Evaluation of systematic characters 181 (I) Abbreviations of names of Institutions 181 IV-Historical sketch 182 V-Definition of the genus 187 VI-Systematic section 188 (a) The variabilis group 188 (i) The variabilis Complex 188 1. G. variabilis 188 2. G. rossica 189 (b) The tibanica group 191 (i) The tibanica Complex 191 3. G. tibanica. 191 4. G. quadrimaculata 192 5. G. ignestii 195 6. G. ornata 196 7. G. trewavasi 198 8. G. makiensis 198 9. G. dembeensis 199 10. G. ethelwynnae 202 (ii) The rufa complex 203 11. G. rufa rufa 203 12. G. rufa obtusa 205 13. O. barteimiae 206 (iii) The lamta complex 208 14. G. lamta 208 15. G. mullya 212 16. G. 'ceylonensis ceylonensis 216 17. G. c. phillipsi 216 18. G. annandalei 217 (173) 174 page (iv) The lissorkynckus complex 219 19. G. lissorkynchus 219 20. G. rupecula 220 ~ (v) The taeniata complex 221 21. G. taeniata. 221 22" G. borneensis 224 (vi) The yunnanensis complex 224 23. G. yunnanensis 225 24. G. gracilis 229 25. G. naganensis 226 26. G. kempii 227 27. G. mcOlellandi 228 28. G. -

Documento Completo Descargar Archivo
Publicaciones científicas del Dr. Raúl A. Ringuelet Zoogeografía y ecología de los peces de aguas continentales de la Argentina y consideraciones sobre las áreas ictiológicas de América del Sur Ecosur, 2(3): 1-122, 1975 Contribución Científica N° 52 al Instituto de Limnología Versión electrónica por: Catalina Julia Saravia (CIC) Instituto de Limnología “Dr. Raúl A. Ringuelet” Enero de 2004 1 Zoogeografía y ecología de los peces de aguas continentales de la Argentina y consideraciones sobre las áreas ictiológicas de América del Sur RAÚL A. RINGUELET SUMMARY: The zoogeography and ecology of fresh water fishes from Argentina and comments on ichthyogeography of South America. This study comprises a critical review of relevant literature on the fish fauna, genocentres, means of dispersal, barriers, ecological groups, coactions, and ecological causality of distribution, including an analysis of allotopic species in the lame lake or pond, the application of indexes of diversity of severa¡ biotopes and comments on historical factors. Its wide scope allows to clarify several aspects of South American Ichthyogeography. The location of Argentina ichthyological fauna according to the above mentioned distributional scheme as well as its relation with the most important hydrography systems are also provided, followed by additional information on its distribution in the Argentine Republic, including an analysis through the application of Simpson's similitude test in several localities. SINOPSIS I. Introducción II. Las hipótesis paleogeográficas de Hermann von Ihering III. La ictiogeografía de Carl H. Eigenmann IV. Estudios de Emiliano J. Mac Donagh sobre distribución de peces argentinos de agua dulce V. El esquema de Pozzi según el patrón hidrográfico actual VI. -

Highly Diversified Late Cretaceous Fish Assemblage Revealed by Otoliths (Ripley Formation and Owl Creek Formation, Northeast Mississippi, Usa)
Rivista Italiana di Paleontologia e Stratigrafia (Research in Paleontology and Stratigraphy) vol. 126(1): 111-155. March 2020 HIGHLY DIVERSIFIED LATE CRETACEOUS FISH ASSEMBLAGE REVEALED BY OTOLITHS (RIPLEY FORMATION AND OWL CREEK FORMATION, NORTHEAST MISSISSIPPI, USA) GARY L. STRINGER1, WERNER SCHWARZHANS*2 , GEORGE PHILLIPS3 & ROGER LAMBERT4 1Museum of Natural History, University of Louisiana at Monroe, Monroe, Louisiana 71209, USA. E-mail: [email protected] 2Natural History Museum of Denmark, Zoological Museum, Universitetsparken 15, DK-2100, Copenhagen, Denmark. E-mail: [email protected] 3Mississippi Museum of Natural Science, 2148 Riverside Drive, Jackson, Mississippi 39202, USA. E-mail: [email protected] 4North Mississippi Gem and Mineral Society, 1817 CR 700, Corinth, Mississippi, 38834, USA. E-mail: [email protected] *Corresponding author To cite this article: Stringer G.L., Schwarzhans W., Phillips G. & Lambert R. (2020) - Highly diversified Late Cretaceous fish assemblage revealed by otoliths (Ripley Formation and Owl Creek Formation, Northeast Mississippi, USA). Riv. It. Paleontol. Strat., 126(1): 111-155. Keywords: Beryciformes; Holocentriformes; Aulopiformes; otolith; evolutionary implications; paleoecology. Abstract. Bulk sampling and extensive, systematic surface collecting of the Coon Creek Member of the Ripley Formation (early Maastrichtian) at the Blue Springs locality and primarily bulk sampling of the Owl Creek Formation (late Maastrichtian) at the Owl Creek type locality, both in northeast Mississippi, USA, have produced the largest and most highly diversified actinopterygian otolith (ear stone) assemblage described from the Mesozoic of North America. The 3,802 otoliths represent 30 taxa of bony fishes representing at least 22 families. In addition, there were two different morphological types of lapilli, which were not identifiable to species level. -

Evolution of Fluvial Drainage During Mountain Building in the Eastern Cordillera of the Colombian Andes
ADVERTIMENT. Lʼaccés als continguts dʼaquesta tesi queda condicionat a lʼacceptació de les condicions dʼús establertes per la següent llicència Creative Commons: http://cat.creativecommons.org/?page_id=184 ADVERTENCIA. El acceso a los contenidos de esta tesis queda condicionado a la aceptación de las condiciones de uso establecidas por la siguiente licencia Creative Commons: http://es.creativecommons.org/blog/licencias/ WARNING. The access to the contents of this doctoral thesis it is limited to the acceptance of the use conditions set by the following Creative Commons license: https://creativecommons.org/licenses/?lang=en Department de Geologia Evolution of fluvial drainage during mountain building in the Eastern Cordillera of the Colombian Andes Ph. D. Thesis 2016 Lucía Struth Izquierdo 1 2 DRAINAGE REORGANIZATION DURING MOUNTAIN BUILDING IN THE EASTERN CORDILLERA OF THE COLOMBIAN ANDES by LUCÍA STRUTH IZQUIERDO A Thesis Submitted in Fulfillment of the Requirements for the Degree of DOCTOR IN GEOLOGY by the UNIVERSITAT AUTÒNOMA DE BARCELONA PhD thesis supervised by Dr. Antonio Teixell Cácharo Departament de Geologia Universitat Autònoma de Barcelona June 2016 3 4 A mi familia. “Talk about a dream, try to make it real” Bruce Springsteen 5 6 Table of contents Abstract ..................................................................................................................................................... 9 Resumen ................................................................................................................................................. -

Amazon Alive!
Amazon Alive! A decade of discovery 1999-2009 The Amazon is the planet’s largest rainforest and river basin. It supports countless thousands of species, as well as 30 million people. © Brent Stirton / Getty Images / WWF-UK © Brent Stirton / Getty Images The Amazon is the largest rainforest on Earth. It’s famed for its unrivalled biological diversity, with wildlife that includes jaguars, river dolphins, manatees, giant otters, capybaras, harpy eagles, anacondas and piranhas. The many unique habitats in this globally significant region conceal a wealth of hidden species, which scientists continue to discover at an incredible rate. Between 1999 and 2009, at least 1,200 new species of plants and vertebrates have been discovered in the Amazon biome (see page 6 for a map showing the extent of the region that this spans). The new species include 637 plants, 257 fish, 216 amphibians, 55 reptiles, 16 birds and 39 mammals. In addition, thousands of new invertebrate species have been uncovered. Owing to the sheer number of the latter, these are not covered in detail by this report. This report has tried to be comprehensive in its listing of new plants and vertebrates described from the Amazon biome in the last decade. But for the largest groups of life on Earth, such as invertebrates, such lists do not exist – so the number of new species presented here is no doubt an underestimate. Cover image: Ranitomeya benedicta, new poison frog species © Evan Twomey amazon alive! i a decade of discovery 1999-2009 1 Ahmed Djoghlaf, Executive Secretary, Foreword Convention on Biological Diversity The vital importance of the Amazon rainforest is very basic work on the natural history of the well known.