MAPK Activation and HRAS Mutation Identified in Pituitary Spindle Cell Oncocytoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

P21 Restrains Pituitary Tumor Growth
p21Cip1 restrains pituitary tumor growth Vera Chesnokovaa, Svetlana Zonisa, Kalman Kovacsb, Anat Ben-Shlomoa, Kolja Wawrowskya, Serguei Bannykhc and Shlomo Melmeda,1 Departments of aMedicine and cPathology, Cedars-Sinai Medical Center, Los Angeles, California 90048, bSt. Michael’s Hospital, Toronto, ON, Canada M5B 1W8 Edited by Wylie W. Vale, The Salk Institute for Biological Studies, La Jolla, CA, and approved September 15, 2008 (received for review May 16, 2008) As commonly encountered, pituitary adenomas are invariably benign. highlighting the securin requirement for the maintenance of chro- We therefore studied protective pituitary proliferative mechanisms. mosomal stability after genotoxic stress (24). Pituitary tumor transforming gene (Pttg) deletion results in pituitary Pituitary-directed transgenic Pttg overexpression results in focal p21 induction and abrogates tumor development in Rb؉/؊Pttg؊/؊ pituitary hyperplasia and small but functional adenoma formation mice. p21 disruption restores attenuated Rb؉/؊Pttg؊/؊ pituitary pro- (14). In contrast, mice lacking Pttg exhibit selective endocrine cell liferation rates and enables high penetrance of pituitary, but not hypoplasia (25). The permissive role of PTTG for pituitary ade- thyroid, tumor growth in triple mutant animals (88% of Rb؉/؊ and noma development was further confirmed by the observation that ϩ Ϫ -of Rb؉/؊Pttg؊/؊p21؊/؊ vs. 30% of Rb؉/؊Pttg؊/؊ mice developed Pttg deletion from the Rb / background results in p53/p21 senes 72% pituitary tumors, P < 0.001). p21 deletion also accelerated S-phase cence pathway activation, associated with pituitary hypoplasia and entry and enhanced transformation rates in triple mutant MEFs. attenuated adenoma formation (23, 26). Intranuclear p21 accumulates in Pttg-null aneuploid GH-secreting p21Cip/Kip (p21), a transcriptional target of p53, is induced in cells, and GH3 rat pituitary tumor cells overexpressing PTTG also response to a spectrum of cellular stresses and acts to constrain the exhibited increased levels of mRNA for both p21 (18-fold, P < 0.01) cell cycle. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

To View the ESE Recommended Curriculum of Specialisation in Clinical Endocrinology, Diabetes and Metabolism
European Society of Endocrinology Recommended Curriculum of Specialisation in Clinical Endocrinology, Diabetes and Metabolism Version 2, November 2019 Contents Endorsement ........................................................................................................................................ 2 Introduction .......................................................................................................................................... 3 1. Diabetes mellitus .............................................................................................................................. 4 2. Lipid disorders ................................................................................................................................... 5 3. Obesity and bariatric endocrinology ................................................................................................. 5 4. Pituitary ............................................................................................................................................ 5 5. Thyroid .............................................................................................................................................. 6 6. Parathyroid, calcium and bone ......................................................................................................... 7 7. Adrenal ............................................................................................................................................. 8 8. Reproductive endocrinology and sexual function -

The Immuno-Neuroendocrine Interface
Amendment history: Erratum (December 2001) Series Introduction: The immuno-neuroendocrine interface Shlomo Melmed J Clin Invest. 2001;108(11):1563-1566. https://doi.org/10.1172/JCI14604. Perspective The CNS and the various endocrine organs are linked in a dynamic manner through networks of reciprocal interactions that allow for both long-term adaptation and short-term shifts in environmental conditions. These regulatory systems embrace the hypothalamus, the pituitary gland, and any of several target glands, including the adrenal gland and the thyroid. Because the anterior pituitary gland secretes at least six key hormones — adrenocorticotropin (ACTH), growth hormone (GH), prolactin (PRL), thyrotropin (TSH), and the gonadotropins follicle stimulating hormone and luteinizing hormone — such diverse parameters as cell integrity, stress responses, growth, development, reproduction, and energy homeostasis are all directly or indirectly under central control (1). In addition, largely because of the dominant role of the hypothalamo-pituitary-adrenal (HPA) axis, inflammation and immunity are also subject to neuroendocrine effects. The articles in this Perspective series explore some of the developmental, physiological, and molecular interactions occurring at the immuno-neuroendocrine interface. Control of pituitary function Three tiers of control subserve regulation of anterior pituitary trophic hormone secretion (2). First, the hypothalamus synthesizes and secretes releasing and inhibiting hormones, which traverse the hypothalamo-pituitary portal system and bind to specific anterior pituitary G protein−linked transmembrane […] Find the latest version: https://jci.me/14604/pdf PERSPECTIVE SERIES Neuro-immune interface Shlomo Melmed, Series Editor SERIES INTRODUCTION The immuno-neuroendocrine interface Shlomo Melmed Cedars-Sinai Research Institute, University of California Los Angeles, School of Medicine, Los Angeles, California 90048, USA. -

RD-Action Matchmaker – Summary of Disease Expertise Recorded Under
Summary of disease expertise recorded via RD-ACTION Matchmaker under each Thematic Grouping and EURORDIS Members’ Thematic Grouping Thematic Reported expertise of those completing the EURORDIS Member perspectives on Grouping matchmaker under each heading Grouping RD Thematically Rare Bone Achondroplasia/Hypochondroplasia Achondroplasia Amelia skeletal dysplasia’s including Achondroplasia/Growth hormone cleidocranial dysostosis, arthrogryposis deficiency/MPS/Turner Brachydactyly chondrodysplasia punctate Fibrous dysplasia of bone Collagenopathy and oncologic disease such as Fibrodysplasia ossificans progressive Li-Fraumeni syndrome Osteogenesis imperfecta Congenital hand and fore-foot conditions Sterno Costo Clavicular Hyperostosis Disorders of Sex Development Duchenne Muscular Dystrophy Ehlers –Danlos syndrome Fibrodysplasia Ossificans Progressiva Growth disorders Hypoparathyroidism Hypophosphatemic rickets & Nutritional Rickets Hypophosphatasia Jeune’s syndrome Limb reduction defects Madelung disease Metabolic Osteoporosis Multiple Hereditary Exostoses Osteogenesis imperfecta Osteoporosis Paediatric Osteoporosis Paget’s disease Phocomelia Pseudohypoparathyroidism Radial dysplasia Skeletal dysplasia Thanatophoric dwarfism Ulna dysplasia Rare Cancer and Adrenocortical tumours Acute monoblastic leukaemia Tumours Carcinoid tumours Brain tumour Craniopharyngioma Colon cancer, familial nonpolyposis Embryonal tumours of CNS Craniopharyngioma Ependymoma Desmoid disease Epithelial thymic tumours in -

Multiple Orbital Neurofibromas, Painful Peripheral Nerve Tumors, Distinctive
European Journal of Human Genetics (2012) 20, 618–625 & 2012 Macmillan Publishers Limited All rights reserved 1018-4813/12 www.nature.com/ejhg ARTICLE Multiple orbital neurofibromas, painful peripheral nerve tumors, distinctive face and marfanoid habitus: a new syndrome D Babovic-Vuksanovic*,1, Ludwine Messiaen2, Christoph Nagel3, Hilde Brems4, Bernd Scheithauer5, Ellen Denayer4, Rong Mao6, Raf Sciot7, Karen M Janowski2, Martin U Schuhmann3, Kathleen Claes8, Eline Beert4, James A Garrity9, Robert J Spinner10, Anat Stemmer-Rachamimov11, Ralitza Gavrilova1, Frank Van Calenbergh12, Victor Mautner13 and Eric Legius4 Four unrelated patients having an unusual clinical phenotype, including multiple peripheral nerve sheath tumors, are reported. Their clinical features were not typical of any known familial tumor syndrome. The patients had multiple painful neurofibromas, including bilateral orbital plexiform neurofibromas, and spinal as well as mucosal neurofibromas. In addition, they exhibited a marfanoid habitus, shared similar facial features, and had enlarged corneal nerves as well as neuronal migration defects. Comprehensive NF1, NF2 and SMARCB1 mutation analyses revealed no mutation in blood lymphocytes and in schwann cells cultured from plexiform neurofibromas. Furthermore, no mutations in RET, PRKAR1A, PTEN and other RAS-pathway genes were found in blood leukocytes. Collectively, the clinical and pathological findings in these four cases fit no known syndrome and likely represent a new disorder. European Journal of Human Genetics (2012) 20, 618–625; doi:10.1038/ejhg.2011.275; published online 18 January 2012 Keywords: orbital neurofibroma; plexiform neurofibroma; enlarged corneal nerves; marfanoid habitus INTRODUCTION Ruvalcaba syndrome, a part of the PHTS spectrum, develop cafe´- Peripheral nerve sheath tumors, specifically neurofibromas and au-lait spots and macules of the glans penis. -

Histochemical Characterization and Functional Significance of the Hyperintense Signal on MR Images of the Posterior Pituitary
1079 Histochemical Characterization and Functional Significance of the Hyperintense Signal on MR Images of the Posterior Pituitary 1 John Kucharczyk ,2 MR imaging of the pituitary fossa characteristically shows a well-circumscribed area Walter Kucharczyk3 of high signal intensity in the posterior lobe on T1-weighted images. We used a Isabelle Berri,4 combination of high-field MR, electron microscopy, and histologic techniques in experi Jack de GroatS mental animals to determine whether the hyperintensity of the posterior lobe might be William Kelly6 functionally related to hormone neurosecretory processes, and to attempt to establish its chemical nature. Histologic sections of a dog's pituitary gland processed with lipid David Norman 1 1 specific markers showed intense staining in the posterior lobe but not in the anterior T. H. Newton lobe, thus documenting the location of fat in the posterior pituitary. Administration of vasoactive drugs known to influence vasopressin secretion to anesthetized cats pro duced changes in the volume of high-intensity signal in the posterior pituitary. Subse quent electron microscopy showed a significant increase in posterior lobe glial cell lipid droplets and neurosecretory granules in dehydration-stimulated cats. The data suggest that the pituitary hyperintensity represents intracellular lipid signal in the glial cell pituicytes of the posterior lobe or neurosecretory granules containing vasopressin. The volume of the signal may, in turn, reflect the functional state of hormonal release from the neurohypophysis. While CT and MR imaging can both be used to evaluate patients with suspected pituitary disease, high-field high-resolution MR imaging is increasingly the method of choice [1-3]. -
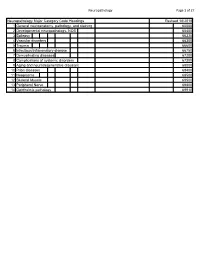
Neuropathology Category Code List
Neuropathology Page 1 of 27 Neuropathology Major Category Code Headings Revised 10/2018 1 General neuroanatomy, pathology, and staining 65000 2 Developmental neuropathology, NOS 65400 3 Epilepsy 66230 4 Vascular disorders 66300 5 Trauma 66600 6 Infectious/inflammatory disease 66750 7 Demyelinating diseases 67200 8 Complications of systemic disorders 67300 9 Aging and neurodegenerative diseases 68000 10 Prion diseases 68400 11 Neoplasms 68500 12 Skeletal Muscle 69500 13 Peripheral Nerve 69800 14 Ophthalmic pathology 69910 Neuropathology Page 2 of 27 Neuropathology 1 General neuroanatomy, pathology, and staining 65000 A Neuroanatomy, NOS 65010 1 Neocortex 65011 2 White matter 65012 3 Entorhinal cortex/hippocampus 65013 4 Deep (basal) nuclei 65014 5 Brain stem 65015 6 Cerebellum 65016 7 Spinal cord 65017 8 Pituitary 65018 9 Pineal 65019 10 Tracts 65020 11 Vascular supply 65021 12 Notochord 65022 B Cell types 65030 1 Neurons 65031 2 Astrocytes 65032 3 Oligodendroglia 65033 4 Ependyma 65034 5 Microglia and mononuclear cells 65035 6 Choroid plexus 65036 7 Meninges 65037 8 Blood vessels 65038 C Cerebrospinal fluid 65045 D Pathologic responses in neurons and axons 65050 1 Axonal degeneration/spheroid/reaction 65051 2 Central chromatolysis 65052 3 Tract degeneration 65053 4 Swollen/ballooned neurons 65054 5 Trans-synaptic neuronal degeneration 65055 6 Olivary hypertrophy 65056 7 Acute ischemic (hypoxic) cell change 65057 8 Apoptosis 65058 9 Protein aggregation 65059 10 Protein degradation/ubiquitin pathway 65060 E Neuronal nuclear inclusions 65100 -

Identification of HRAS Mutations and Absence of GNAQ Or GNA11
Modern Pathology (2013) 26, 1320–1328 1320 & 2013 USCAP, Inc All rights reserved 0893-3952/13 $32.00 Identification of HRAS mutations and absence of GNAQ or GNA11 mutations in deep penetrating nevi Ryan P Bender1, Matthew J McGinniss2, Paula Esmay1, Elsa F Velazquez3,4 and Julie DR Reimann3,4 1Caris Life Sciences, Phoenix, AZ, USA; 2Genoptix Medical Laboratory, Carlsbad, CA, USA; 3Dermatopathology Division, Miraca Life Sciences Research Institute, Newton, MA, USA and 4Department of Dermatology, Tufts Medical Center, Boston, MA, USA HRAS is mutated in B15% of Spitz nevi, and GNAQ or GNA11 is mutated in blue nevi (46–83% and B7% respectively). Epithelioid blue nevi and deep penetrating nevi show features of both blue nevi (intradermal location, pigmentation) and Spitz nevi (epithelioid morphology). Epithelioid blue nevi and deep penetrating nevi can also show overlapping features with melanoma, posing a diagnostic challenge. Although epithelioid blue nevi are considered blue nevic variants, no GNAQ or GNA11 mutations have been reported. Classification of deep penetrating nevi as blue nevic variants has also been proposed, however, no GNAQ or GNA11 mutations have been reported and none have been tested for HRAS mutations. To better characterize these tumors, we performed mutational analysis for GNAQ, GNA11, and HRAS, with blue nevi and Spitz nevi as controls. Within deep penetrating nevi, none demonstrated GNAQ or GNA11 mutations (0/38). However, 6% revealed HRAS mutation (2/32). Twenty percent of epithelioid blue nevi contained a GNAQ mutation (2/10), while none displayed GNA11 or HRAS mutation. Eighty-seven percent of blue nevi contained a GNAQ mutation (26/30), 4% a GNA11 mutation (1/28), and none an HRAS mutation. -

Role of Phosphodiesterases in the Pathophysiology of Neurodevelopmental Disorders
Molecular Psychiatry https://doi.org/10.1038/s41380-020-00997-9 REVIEW ARTICLE Role of phosphodiesterases in the pathophysiology of neurodevelopmental disorders 1 2 Sébastien Delhaye ● Barbara Bardoni Received: 4 July 2020 / Revised: 3 December 2020 / Accepted: 9 December 2020 © The Author(s), under exclusive licence to Springer Nature Limited part of Springer Nature 2021. This article is published with open access Abstract Phosphodiesterases (PDEs) are enzymes involved in the homeostasis of both cAMP and cGMP. They are members of a family of proteins that includes 11 subfamilies with different substrate specificities. Their main function is to catalyze the hydrolysis of cAMP, cGMP, or both. cAMP and cGMP are two key second messengers that modulate a wide array of intracellular processes and neurobehavioral functions, including memory and cognition. Even if these enzymes are present in all tissues, we focused on those PDEs that are expressed in the brain. We took into consideration genetic variants in patients affected by neurodevelopmental disorders, phenotypes of animal models, and pharmacological effects of PDE inhibitors, a class of drugs in rapid evolution and increasing application to brain disorders. Collectively, these data indicate the potential 1234567890();,: 1234567890();,: of PDE modulators to treat neurodevelopmental diseases characterized by learning and memory impairment, alteration of behaviors associated with depression, and deficits in social interaction. Indeed, clinical trials are in progress to treat patients with Alzheimer’s disease, schizophrenia, depression, and autism spectrum disorders. Among the most recent results, the application of some PDE inhibitors (PDE2A, PDE3, PDE4/4D, and PDE10A) to treat neurodevelopmental diseases, including autism spectrum disorders and intellectual disability, is a significant advance, since no specific therapies are available for these disorders that have a large prevalence. -

Prevalence of Mutations in TSHR, GNAS, PRKAR1A and RAS Genes
European Journal of Endocrinology (2008) 159 623–631 ISSN 0804-4643 CLINICAL STUDY Prevalence of mutations in TSHR, GNAS, PRKAR1A and RAS genes in a large series of toxic thyroid adenomas from Galicia, an iodine-deficient area in NW Spain F Palos-Paz1, O Perez-Guerra1, J Cameselle-Teijeiro3,CRueda-Chimeno5, F Barreiro-Morandeira4, J Lado-Abeal1,2 and the Galician Group for the Study of Toxic Multinodular Goitre: D Araujo Vilar1,2, R Argueso7, O Barca1, MBotana7, J M Cabezas-Agrı´cola2, P Catalina6, L Dominguez Gerpe1, T Fernandez9, A Mato8, A Nun˜o11,MPenin10 and B Victoria1 1Unidade de Enfermedades Tiroideas e Metabo´licas (UETeM), 2Endocrinology Section, Department of Medicine, 3Pathology Department and 4Surgery Department, Complexo Hospitalario Universitary de Santiago (CHUS), University of Santiago de Compostela, Santiago de Compostela, 15705, Spain, 5General Surgery Section and 6Endocrinology Section, Complexo Hospitalario de Pontevedra, Pontevedra, Spain, 7Endocrinology Section, Complexo Hospitalario Xeral-Calde, Lugo, Spain, 8Endocrinology Section, Complexo Hospitalario de Ourense, Ourense, Spain, 9Endocrinology Section, Complexo Hospitalario Universitario Juan-Canalejo, A Corun˜a, Spain, 10Endocrinology Section, Hospital Arquitecto Marcide, Ferrol, Spain and 11General Surgery Section, Hospital do Meixoeiro, Complexo Hospitalario Universitario de Vigo, Vigo, Spain (Correspondence should be addressed to J Lado-Abeal who is now at UETeM, Department of Medicine, School of Medicine, University of Santiago de Compostela, C/San Francisco sn 15705, Santiago de Compostela, Spain; Email: [email protected]) Abstract Objective: Toxic thyroid adenoma (TA) is a common cause of hyperthyroidism. Mutations in the TSH receptor (TSHR) gene, and less frequently in the adenylate cyclase-stimulating G alpha protein (GNAS) gene, are well established causes of TA in Europe. -

Genomic Alterations Associated with Recurrence and TNBC Subtype in High
Author Manuscript Published OnlineFirst on August 31, 2018; DOI: 10.1158/1541-7786.MCR-18-0619 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Genomic Alterations Associated with Recurrence and TNBC Subtype in High- risk Early Breast Cancers Timothy R. Wilson1*, Akshata R. Udyavar2*, Ching-Wei Chang3, Jill M. Spoerke1, Junko Aimi1, Heidi M. Savage1, Anneleen Daemen2, Joyce A. O’Shaughnessy4,5,6, Richard Bourgon2 and Mark R. Lackner1 Departments of 1Oncology Biomarker Development, 2Bioinformatics and Computational Biology, 3Biostatistics, Genentech Inc., 1 DNA Way, South San Francisco, CA, USA.4US Oncology, 5Baylor University Medical Center, 6Texas Oncology, Dallas, TX, USA. *These authors contributed equally to this work. Correspondence and request for materials should be addressed to T.R.W. (email: [email protected]) or M.R.L. (email: [email protected]). Running Title: Genomic Profiling of High-risk Early Breast Cancers Keywords: early breast cancer, next generation sequencing, risk of recurrence, triple negative breast cancer, tumor mutational burden Conflicts of interest: TRW, ARY, CWC, JMS, JA, HMS, AD, RB and MRL are employed by Genentech and have stocks in Roche. JOS has received speaking fees from Blue Earth Diagnostics and Novartis, consulting fees from AstraZeneca, Celgene, Eli Lilly, LaRoche, Lilly USA, Merck, Novartis and Pfizer, Honoraria from Seattle Genetics and Downloaded from mcr.aacrjournals.org on September 27, 2021. © 2018 American Association for Cancer Research. Author Manuscript Published OnlineFirst on August 31, 2018; DOI: 10.1158/1541-7786.MCR-18-0619 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited.