Exome Sequencing Identifies PDE4D Mutations As Another Cause of Acrodysostosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

To View the ESE Recommended Curriculum of Specialisation in Clinical Endocrinology, Diabetes and Metabolism
European Society of Endocrinology Recommended Curriculum of Specialisation in Clinical Endocrinology, Diabetes and Metabolism Version 2, November 2019 Contents Endorsement ........................................................................................................................................ 2 Introduction .......................................................................................................................................... 3 1. Diabetes mellitus .............................................................................................................................. 4 2. Lipid disorders ................................................................................................................................... 5 3. Obesity and bariatric endocrinology ................................................................................................. 5 4. Pituitary ............................................................................................................................................ 5 5. Thyroid .............................................................................................................................................. 6 6. Parathyroid, calcium and bone ......................................................................................................... 7 7. Adrenal ............................................................................................................................................. 8 8. Reproductive endocrinology and sexual function -

Open Full Page
Research Article Distinct Genomic Profiles in Hereditary Breast Tumors Identified by Array-Based Comparative Genomic Hybridization Go¨ran Jo¨nsson,1 Tara L. Naylor,5 Johan Vallon-Christersson,1 Johan Staaf,1 Jia Huang,5 M. Renee Ward,5 Joel D. Greshock,5 Lena Luts,4 Ha˚kan Olsson,1 Nazneen Rahman,6 Michael Stratton,6 Markus Ringne´r,3 A˚ke Borg,1,2 and Barbara L. Weber5 1Department of Oncology, University Hospital; 2Lund Strategic Research Center for Stem Cell Biology and Cell Therapy and 3Department of Theoretical Physics, Lund University; and 4Department of Pathology, University Hospital, Lund, Sweden; 5Abramson Family Cancer Research Institute, University of Pennsylvania, Philadelphia, Pennsylvania; and 6Section of Cancer Genetics, Institute of Cancer Research, Sutton, Surrey, United Kingdom Abstract of additional predisposing genes, although technical limitations Mutations in BRCA1 and BRCA2 account for a significant and the complexity of BRCA gene regulation and mutation proportion of hereditary breast cancers. Earlier studies have spectrum can probably explain why some disease-causing muta- shown that inherited and sporadic tumors progress along tions are missed (2). BRCA1 and BRCA2 function as classic tumor different somatic genetic pathways and that global gene suppressor genes with frequent loss of the wild-type allele in expression profiles distinguish between these groups. To tumors of mutation carriers. The BRCA1 protein has been determine whether genomic profiles similarly discriminate implicated in a broad range of cellular functions, including repair among BRCA1, BRCA2, and sporadic tumors, we established of double-strand breaks by homologous recombination, cell cycle DNA copy number profiles using comparative genomic checkpoint control, chromatin remodeling, and transcriptional hybridization to BAC-clone microarrays providing <1 Mb regulation. -

RD-Action Matchmaker – Summary of Disease Expertise Recorded Under
Summary of disease expertise recorded via RD-ACTION Matchmaker under each Thematic Grouping and EURORDIS Members’ Thematic Grouping Thematic Reported expertise of those completing the EURORDIS Member perspectives on Grouping matchmaker under each heading Grouping RD Thematically Rare Bone Achondroplasia/Hypochondroplasia Achondroplasia Amelia skeletal dysplasia’s including Achondroplasia/Growth hormone cleidocranial dysostosis, arthrogryposis deficiency/MPS/Turner Brachydactyly chondrodysplasia punctate Fibrous dysplasia of bone Collagenopathy and oncologic disease such as Fibrodysplasia ossificans progressive Li-Fraumeni syndrome Osteogenesis imperfecta Congenital hand and fore-foot conditions Sterno Costo Clavicular Hyperostosis Disorders of Sex Development Duchenne Muscular Dystrophy Ehlers –Danlos syndrome Fibrodysplasia Ossificans Progressiva Growth disorders Hypoparathyroidism Hypophosphatemic rickets & Nutritional Rickets Hypophosphatasia Jeune’s syndrome Limb reduction defects Madelung disease Metabolic Osteoporosis Multiple Hereditary Exostoses Osteogenesis imperfecta Osteoporosis Paediatric Osteoporosis Paget’s disease Phocomelia Pseudohypoparathyroidism Radial dysplasia Skeletal dysplasia Thanatophoric dwarfism Ulna dysplasia Rare Cancer and Adrenocortical tumours Acute monoblastic leukaemia Tumours Carcinoid tumours Brain tumour Craniopharyngioma Colon cancer, familial nonpolyposis Embryonal tumours of CNS Craniopharyngioma Ependymoma Desmoid disease Epithelial thymic tumours in -

Multiple Orbital Neurofibromas, Painful Peripheral Nerve Tumors, Distinctive
European Journal of Human Genetics (2012) 20, 618–625 & 2012 Macmillan Publishers Limited All rights reserved 1018-4813/12 www.nature.com/ejhg ARTICLE Multiple orbital neurofibromas, painful peripheral nerve tumors, distinctive face and marfanoid habitus: a new syndrome D Babovic-Vuksanovic*,1, Ludwine Messiaen2, Christoph Nagel3, Hilde Brems4, Bernd Scheithauer5, Ellen Denayer4, Rong Mao6, Raf Sciot7, Karen M Janowski2, Martin U Schuhmann3, Kathleen Claes8, Eline Beert4, James A Garrity9, Robert J Spinner10, Anat Stemmer-Rachamimov11, Ralitza Gavrilova1, Frank Van Calenbergh12, Victor Mautner13 and Eric Legius4 Four unrelated patients having an unusual clinical phenotype, including multiple peripheral nerve sheath tumors, are reported. Their clinical features were not typical of any known familial tumor syndrome. The patients had multiple painful neurofibromas, including bilateral orbital plexiform neurofibromas, and spinal as well as mucosal neurofibromas. In addition, they exhibited a marfanoid habitus, shared similar facial features, and had enlarged corneal nerves as well as neuronal migration defects. Comprehensive NF1, NF2 and SMARCB1 mutation analyses revealed no mutation in blood lymphocytes and in schwann cells cultured from plexiform neurofibromas. Furthermore, no mutations in RET, PRKAR1A, PTEN and other RAS-pathway genes were found in blood leukocytes. Collectively, the clinical and pathological findings in these four cases fit no known syndrome and likely represent a new disorder. European Journal of Human Genetics (2012) 20, 618–625; doi:10.1038/ejhg.2011.275; published online 18 January 2012 Keywords: orbital neurofibroma; plexiform neurofibroma; enlarged corneal nerves; marfanoid habitus INTRODUCTION Ruvalcaba syndrome, a part of the PHTS spectrum, develop cafe´- Peripheral nerve sheath tumors, specifically neurofibromas and au-lait spots and macules of the glans penis. -

Polyclonal Antibody to PDE4D - Aff - Purified
OriGene Technologies, Inc. OriGene Technologies GmbH 9620 Medical Center Drive, Ste 200 Schillerstr. 5 Rockville, MD 20850 32052 Herford UNITED STATES GERMANY Phone: +1-888-267-4436 Phone: +49-5221-34606-0 Fax: +1-301-340-8606 Fax: +49-5221-34606-11 [email protected] [email protected] AP06558PU-N Polyclonal Antibody to PDE4D - Aff - Purified Alternate names: 5'-cyclic phosphodiesterase 4D, DPDE3, DPDE3, EC=3.1.4.17, PDE43, PDE4D, cAMP- specific 3' Quantity: 0.1 mg Concentration: 1.0 mg/ml Background: Phosphodiesterases (PDE) hydrolyze cAMP to 5’AMP and thus play a critical role in the regulation of intracellular cAMP. Division of the PDE superfamily by sequence homology and enzymatic properties yields 11 PDE families. A unique upstream conserved region (UCR) characterizes the PDE4 family. Four separate genes (A-D) encode the PDE4 enzymes, and alternative splicing generates short or long isoforms of each gene. Long PDE4 isoforms contain both UCR1 and UCR2 while short PDE4 isoforms possess only UCR2. Both UCR domains are necessary for dimerization of PDE4 isoforms. The human PDE4D gene maps to chromosome 5q12. The splice variants include isoforms PDE4D1-6. Uniprot ID: Q08499 NCBI: NP_001098101.1 GeneID: 5144 Host: Rabbit Immunogen: Synthetic peptide, corresponding to amino acids 30-70 Human PDE4D. Format: State: Liquid purified Ig fraction Purification: Affinity-chromatography using epitope-specific immunogen and the purity is > 95% (by SDS-PAGE) Buffer System: Phosphate buffered saline (PBS), pH 7.2. Preservatives: 15 mM sodium azide Applications: Western blot: 1/500-1/1000. Immunohistochemistry on paraffin sections: 1/50-1/200. -
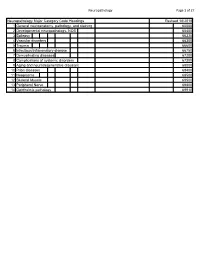
Neuropathology Category Code List
Neuropathology Page 1 of 27 Neuropathology Major Category Code Headings Revised 10/2018 1 General neuroanatomy, pathology, and staining 65000 2 Developmental neuropathology, NOS 65400 3 Epilepsy 66230 4 Vascular disorders 66300 5 Trauma 66600 6 Infectious/inflammatory disease 66750 7 Demyelinating diseases 67200 8 Complications of systemic disorders 67300 9 Aging and neurodegenerative diseases 68000 10 Prion diseases 68400 11 Neoplasms 68500 12 Skeletal Muscle 69500 13 Peripheral Nerve 69800 14 Ophthalmic pathology 69910 Neuropathology Page 2 of 27 Neuropathology 1 General neuroanatomy, pathology, and staining 65000 A Neuroanatomy, NOS 65010 1 Neocortex 65011 2 White matter 65012 3 Entorhinal cortex/hippocampus 65013 4 Deep (basal) nuclei 65014 5 Brain stem 65015 6 Cerebellum 65016 7 Spinal cord 65017 8 Pituitary 65018 9 Pineal 65019 10 Tracts 65020 11 Vascular supply 65021 12 Notochord 65022 B Cell types 65030 1 Neurons 65031 2 Astrocytes 65032 3 Oligodendroglia 65033 4 Ependyma 65034 5 Microglia and mononuclear cells 65035 6 Choroid plexus 65036 7 Meninges 65037 8 Blood vessels 65038 C Cerebrospinal fluid 65045 D Pathologic responses in neurons and axons 65050 1 Axonal degeneration/spheroid/reaction 65051 2 Central chromatolysis 65052 3 Tract degeneration 65053 4 Swollen/ballooned neurons 65054 5 Trans-synaptic neuronal degeneration 65055 6 Olivary hypertrophy 65056 7 Acute ischemic (hypoxic) cell change 65057 8 Apoptosis 65058 9 Protein aggregation 65059 10 Protein degradation/ubiquitin pathway 65060 E Neuronal nuclear inclusions 65100 -

Analysis of the Indacaterol-Regulated Transcriptome in Human Airway
Supplemental material to this article can be found at: http://jpet.aspetjournals.org/content/suppl/2018/04/13/jpet.118.249292.DC1 1521-0103/366/1/220–236$35.00 https://doi.org/10.1124/jpet.118.249292 THE JOURNAL OF PHARMACOLOGY AND EXPERIMENTAL THERAPEUTICS J Pharmacol Exp Ther 366:220–236, July 2018 Copyright ª 2018 by The American Society for Pharmacology and Experimental Therapeutics Analysis of the Indacaterol-Regulated Transcriptome in Human Airway Epithelial Cells Implicates Gene Expression Changes in the s Adverse and Therapeutic Effects of b2-Adrenoceptor Agonists Dong Yan, Omar Hamed, Taruna Joshi,1 Mahmoud M. Mostafa, Kyla C. Jamieson, Radhika Joshi, Robert Newton, and Mark A. Giembycz Departments of Physiology and Pharmacology (D.Y., O.H., T.J., K.C.J., R.J., M.A.G.) and Cell Biology and Anatomy (M.M.M., R.N.), Snyder Institute for Chronic Diseases, Cumming School of Medicine, University of Calgary, Calgary, Alberta, Canada Received March 22, 2018; accepted April 11, 2018 Downloaded from ABSTRACT The contribution of gene expression changes to the adverse and activity, and positive regulation of neutrophil chemotaxis. The therapeutic effects of b2-adrenoceptor agonists in asthma was general enriched GO term extracellular space was also associ- investigated using human airway epithelial cells as a therapeu- ated with indacaterol-induced genes, and many of those, in- tically relevant target. Operational model-fitting established that cluding CRISPLD2, DMBT1, GAS1, and SOCS3, have putative jpet.aspetjournals.org the long-acting b2-adrenoceptor agonists (LABA) indacaterol, anti-inflammatory, antibacterial, and/or antiviral activity. Numer- salmeterol, formoterol, and picumeterol were full agonists on ous indacaterol-regulated genes were also induced or repressed BEAS-2B cells transfected with a cAMP-response element in BEAS-2B cells and human primary bronchial epithelial cells by reporter but differed in efficacy (indacaterol $ formoterol . -

Identification of HRAS Mutations and Absence of GNAQ Or GNA11
Modern Pathology (2013) 26, 1320–1328 1320 & 2013 USCAP, Inc All rights reserved 0893-3952/13 $32.00 Identification of HRAS mutations and absence of GNAQ or GNA11 mutations in deep penetrating nevi Ryan P Bender1, Matthew J McGinniss2, Paula Esmay1, Elsa F Velazquez3,4 and Julie DR Reimann3,4 1Caris Life Sciences, Phoenix, AZ, USA; 2Genoptix Medical Laboratory, Carlsbad, CA, USA; 3Dermatopathology Division, Miraca Life Sciences Research Institute, Newton, MA, USA and 4Department of Dermatology, Tufts Medical Center, Boston, MA, USA HRAS is mutated in B15% of Spitz nevi, and GNAQ or GNA11 is mutated in blue nevi (46–83% and B7% respectively). Epithelioid blue nevi and deep penetrating nevi show features of both blue nevi (intradermal location, pigmentation) and Spitz nevi (epithelioid morphology). Epithelioid blue nevi and deep penetrating nevi can also show overlapping features with melanoma, posing a diagnostic challenge. Although epithelioid blue nevi are considered blue nevic variants, no GNAQ or GNA11 mutations have been reported. Classification of deep penetrating nevi as blue nevic variants has also been proposed, however, no GNAQ or GNA11 mutations have been reported and none have been tested for HRAS mutations. To better characterize these tumors, we performed mutational analysis for GNAQ, GNA11, and HRAS, with blue nevi and Spitz nevi as controls. Within deep penetrating nevi, none demonstrated GNAQ or GNA11 mutations (0/38). However, 6% revealed HRAS mutation (2/32). Twenty percent of epithelioid blue nevi contained a GNAQ mutation (2/10), while none displayed GNA11 or HRAS mutation. Eighty-seven percent of blue nevi contained a GNAQ mutation (26/30), 4% a GNA11 mutation (1/28), and none an HRAS mutation. -

Melanomas Are Comprised of Multiple Biologically Distinct Categories
Melanomas are comprised of multiple biologically distinct categories, which differ in cell of origin, age of onset, clinical and histologic presentation, pattern of metastasis, ethnic distribution, causative role of UV radiation, predisposing germ line alterations, mutational processes, and patterns of somatic mutations. Neoplasms are initiated by gain of function mutations in one of several primary oncogenes, typically leading to benign melanocytic nevi with characteristic histologic features. The progression of nevi is restrained by multiple tumor suppressive mechanisms. Secondary genetic alterations override these barriers and promote intermediate or overtly malignant tumors along distinct progression trajectories. The current knowledge about pathogenesis, clinical, histological and genetic features of primary melanocytic neoplasms is reviewed and integrated into a taxonomic framework. THE MOLECULAR PATHOLOGY OF MELANOMA: AN INTEGRATED TAXONOMY OF MELANOCYTIC NEOPLASIA Boris C. Bastian Corresponding Author: Boris C. Bastian, M.D. Ph.D. Gerson & Barbara Bass Bakar Distinguished Professor of Cancer Biology Departments of Dermatology and Pathology University of California, San Francisco UCSF Cardiovascular Research Institute 555 Mission Bay Blvd South Box 3118, Room 252K San Francisco, CA 94158-9001 [email protected] Key words: Genetics Pathogenesis Classification Mutation Nevi Table of Contents Molecular pathogenesis of melanocytic neoplasia .................................................... 1 Classification of melanocytic neoplasms -

Blueprint Genetics Comprehensive Growth Disorders / Skeletal
Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel Test code: MA4301 Is a 374 gene panel that includes assessment of non-coding variants. This panel covers the majority of the genes listed in the Nosology 2015 (PMID: 26394607) and all genes in our Malformation category that cause growth retardation, short stature or skeletal dysplasia and is therefore a powerful diagnostic tool. It is ideal for patients suspected to have a syndromic or an isolated growth disorder or a skeletal dysplasia. About Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders This panel covers a broad spectrum of diseases associated with growth retardation, short stature or skeletal dysplasia. Many of these conditions have overlapping features which can make clinical diagnosis a challenge. Genetic diagnostics is therefore the most efficient way to subtype the diseases and enable individualized treatment and management decisions. Moreover, detection of causative mutations establishes the mode of inheritance in the family which is essential for informed genetic counseling. For additional information regarding the conditions tested on this panel, please refer to the National Organization for Rare Disorders and / or GeneReviews. Availability 4 weeks Gene Set Description Genes in the Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ACAN# Spondyloepimetaphyseal dysplasia, aggrecan type, AD/AR 20 56 Spondyloepiphyseal dysplasia, Kimberley -

Blueprint Genetics Comprehensive Skeletal Dysplasias and Disorders
Comprehensive Skeletal Dysplasias and Disorders Panel Test code: MA3301 Is a 251 gene panel that includes assessment of non-coding variants. Is ideal for patients with a clinical suspicion of disorders involving the skeletal system. About Comprehensive Skeletal Dysplasias and Disorders This panel covers a broad spectrum of skeletal disorders including common and rare skeletal dysplasias (eg. achondroplasia, COL2A1 related dysplasias, diastrophic dysplasia, various types of spondylo-metaphyseal dysplasias), various ciliopathies with skeletal involvement (eg. short rib-polydactylies, asphyxiating thoracic dysplasia dysplasias and Ellis-van Creveld syndrome), various subtypes of osteogenesis imperfecta, campomelic dysplasia, slender bone dysplasias, dysplasias with multiple joint dislocations, chondrodysplasia punctata group of disorders, neonatal osteosclerotic dysplasias, osteopetrosis and related disorders, abnormal mineralization group of disorders (eg hypopohosphatasia), osteolysis group of disorders, disorders with disorganized development of skeletal components, overgrowth syndromes with skeletal involvement, craniosynostosis syndromes, dysostoses with predominant craniofacial involvement, dysostoses with predominant vertebral involvement, patellar dysostoses, brachydactylies, some disorders with limb hypoplasia-reduction defects, ectrodactyly with and without other manifestations, polydactyly-syndactyly-triphalangism group of disorders, and disorders with defects in joint formation and synostoses. Availability 4 weeks Gene Set Description -

Hypotyreose V01
2/18/2021 Medfødt hypotyreose v01 Avdeling for medisinsk genetikk Medfødt hypotyreose Genpanel, versjon v01 * Enkelte genomiske regioner har lav eller ingen sekvensdekning ved eksomsekvensering. Dette skyldes at de har stor likhet med andre områder i genomet, slik at spesifikk gjenkjennelse av disse områdene og påvisning av varianter i disse områdene, blir vanskelig og upålitelig. Disse genetiske regionene har vi identifisert ved å benytte USCS segmental duplication hvor områder større enn 1 kb og ≥90% likhet med andre regioner i genomet, gjenkjennes (https://genome.ucsc.edu). Vi gjør oppmerksom på at ved identifiseringav ekson oppstrøms for startkodon kan eksonnummereringen endres uten at transkript ID endres. Avdelingens websider har en full oversikt over områder som er affisert av segmentale duplikasjoner. ** Transkriptets kodende ekson. Gen Gen Ekson (HGNC (HGNC Transkript affisert av Ekson** Fenotype symbol) ID) segdup* CDCA8 14629 NM_001256875.2 1-10 DUOX2 13273 NM_014080.4 5-8 2-34 Thyroid dyshormonogenesis 6 OMIM DUOXA2 32698 NM_207581.4 1-6 Thyroid dyshormonogenesis 5 OMIM FOXE1 3806 NM_004473.4 1 Bamforth-Lazarus syndrome OMIM GLIS3 28510 NM_152629.3 2-10 Diabetes mellitus, neonatal, with congenital hypothyroidism OMIM file:///data/Hypotyreose_v01-web.html 1/4 2/18/2021 Medfødt hypotyreose v01 Gen Gen Ekson (HGNC (HGNC Transkript affisert av Ekson** Fenotype symbol) ID) segdup* GNAS 4392 NM_000516.6 1-13 ACTH-independent macronodular adrenal hyperplasia OMIM McCune-Albright syndrome, somatic, mosaic OMIM Osseous heteroplasia, progressive