Extracting Morphometric Information from Rat Sciatic Nerve Using Optical Coherence Tomography
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nerve Ultrasound in Dorsal Root Ganglion Disorders: Smaller Nerves Lead to Bigger Insights
Clinical Neurophysiology 130 (2019) 550–551 Contents lists available at ScienceDirect Clinical Neurophysiology journal homepage: www.elsevier.com/locate/clinph Editorial Nerve ultrasound in dorsal root ganglion disorders: Smaller nerves lead to bigger insights See Article, pages 568–572 After decades of having to make do with electric stimulation representing the fascicles, bundled together in a large outer cable and recording (i.e. nerve conduction studies, electromyography sheath (van Alfen et al., 2018). and evoked potentials), nerve ultrasound now provides the oppor- Next, it is important to realize what the ratio between axon/ tunity to improve neurodiagnostic patient care by deploying a myelin and connective tissue in a given nerve segment is, and powerful tool to detect neuromuscular pathology in an accurate how that ratio changes from the proximal root to the distal end and patient-friendly way (Mah et al., 2018; Walker et al., 2018). branches (Schraut et al., 2016). Connective tissue elements of the Nerve ultrasound is also increasingly providing neurologists and perineurium and epineurium are relatively sparse at the very prox- clinical neurophysiologists with the opportunity to increase their imal root and plexus levels, with an average connective tissue con- insight in the pathophysiology of peripheral nervous system tent of around 25–30%. Ultrasonographically, this means that roots (PNS) pathology. In this issue of Clinical Neurophysiology, Leadbet- will always look rather black in appearance without much dis- ter and coworkers (Leadbetter et al., 2019) describe the results of cernible fascicular architecture, as the sparseness of connective tis- their study on nerve ultrasound for diagnosing sensory neuronopa- sue elements provides relatively few reflectors to create an image thy in spinocerebellar ataxia type 2 and CANVAS syndrome. -
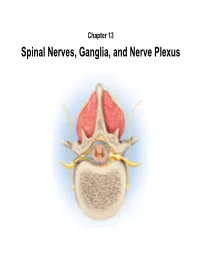
Spinal Nerves, Ganglia, and Nerve Plexus Spinal Nerves
Chapter 13 Spinal Nerves, Ganglia, and Nerve Plexus Spinal Nerves Posterior Spinous process of vertebra Posterior root Deep muscles of back Posterior ramus Spinal cord Transverse process of vertebra Posterior root ganglion Spinal nerve Anterior ramus Meningeal branch Communicating rami Anterior root Vertebral body Sympathetic ganglion Anterior General Anatomy of Nerves and Ganglia • Spinal cord communicates with the rest of the body by way of spinal nerves • nerve = a cordlike organ composed of numerous nerve fibers (axons) bound together by connective tissue – mixed nerves contain both afferent (sensory) and efferent (motor) fibers – composed of thousands of fibers carrying currents in opposite directions Anatomy of a Nerve Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Epineurium Perineurium Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Endoneurium Nerve Rootlets fiber Posterior root Fascicle Posterior root ganglion Anterior Blood root vessels Spinal nerve (b) Copyright by R.G. Kessel and R.H. Kardon, Tissues and Organs: A Text-Atlas of Scanning Electron Microscopy, 1979, W.H. Freeman, All rights reserved Blood vessels Fascicle Epineurium Perineurium Unmyelinated nerve fibers Myelinated nerve fibers (a) Endoneurium Myelin General Anatomy of Nerves and Ganglia • nerves of peripheral nervous system are ensheathed in Schwann cells – forms neurilemma and often a myelin sheath around the axon – external to neurilemma, each fiber is surrounded by -

The Peripheral Nervous System
The Peripheral Nervous System Dr. Ali Ebneshahidi Peripheral Nervous System (PNS) – Consists of 12 pairs of cranial nerves and 31 pairs of spinal nerves. – Serves as a critical link between the body and the central nervous system. – peripheral nerves contain an outermost layer of fibrous connective tissue called epineurium which surrounds a thinner layer of fibrous connective tissue called perineurium (surrounds the bundles of nerve or fascicles). Individual nerve fibers within the nerve are surrounded by loose connective tissue called endoneurium. Cranial Nerves Cranial nerves are direct extensions of the brain. Only Nerve I (olfactory) originates from the cerebrum, the remaining 11 pairs originate from the brain stem. Nerve I (Olfactory)- for the sense of smell (sensory). Nerve II (Optic)- for the sense of vision (sensory). Nerve III (Oculomotor)- for controlling muscles and accessory structures of the eyes ( primarily motor). Nerve IV (Trochlear)- for controlling muscles of the eyes (primarily motor). Nerve V (Trigeminal)- for controlling muscles of the eyes, upper and lower jaws and tear glands (mixed). Nerve VI (Abducens)- for controlling muscles that move the eye (primarily motor). Nerve VII (Facial) – for the sense of taste and controlling facial muscles, tear glands and salivary glands (mixed). Nerve VIII (Vestibulocochlear)- for the senses of hearing and equilibrium (sensory). Nerve IX (Glossopharyngeal)- for controlling muscles in the pharynx and to control salivary glands (mixed). Nerve X (Vagus)- for controlling muscles used in speech, swallowing, and the digestive tract, and controls cardiac and smooth muscles (mixed). Nerve XI (Accessory)- for controlling muscles of soft palate, pharynx and larynx (primarily motor). Nerve XII (Hypoglossal) for controlling muscles that move the tongue ( primarily motor). -
PN1 (Midha) Microanatomy of Peripheral Nerves-Part1.Pdf
Peripheral Nerve Basics • Consist of processes of cell bodies found in the DRG, anterior horn, and autonomic ganglia • Organized by several distinct connective tissue layers – the epineurium, perineurium, and endoneurium • Vascular supply provided by the vasa nervorum Peripheral Nerve Basics • Neuronal processes bound into fascicles by perineurium • Fascicles bound into nerves by epineurium • Endoneurium is a division of the perineurium which form thin layers of connective tissue surrounding neuronal fibers in a fascicle Sural nerve in cross-section Epineurium • Loose areolar tissue with sparse, longitudinally-oriented collagen fibers • Some elastic fibers where epineurium abuts perineurium • Able to accommodate a significant amount of nerve stretching and movement • Increases in thickness where nerves cross joints • Constitutes an increasing proportion of nerves as they increase in size • Epineurial fat helps cushion nerves from compressive injury • Decreased epineurial fat found in patients with diabetes Perineurium • Cellular component composed of laminated fibroblasts of up to 15 layers in thickness which are bounded by a basal lamina • Semi-permeable: inner lamellae have tight junctions, providing a barrier to intercellular transport of macromolecules – Tight junctions can be loosened with topical anaesthetics and with osmotic change Perineurium • Exhibits a slightly positive internal pressure – Fascicular contents herniate upon perineurial injury • Under tension longitudinally – Nerve segment shortens upon transection – may complicate surgical repair as nerve can be stretched only approximately 10% before being inhibited by collagen Endoneurium • Intrafascicular connective tissue consisting of a collagenous matrix in the interstitial space • Develops into partitions of dense connective tissue between diverging fascicles and eventually becomes perineurium when the fascicles separate • Collagen fibers are longitudinally-oriented and run along nerve fibers and capillaries. -

Lecture 19: Nerve Synthesis in Vivo (Regeneration)
Nerve synthesis in vivo (regeneration)* 1. Anatomy and function of a peripheral nerve. 2. Experimental parameters for study of induced regeneration. 3. Synthesis of myelinated axons and BM (nerve fibers) 4. Evidence (?) of synthesis of an endoneurium. 5. Synthesis of a nerve trunk (including summary of kinetics of synthesis). 6. Comparative regenerative activity of various reactants. _______ *Tissue and Organ Regeneration in Adults, Yannas, Springer, 2001, Ch. 6. 1. Anatomy and function of a peripheral nerve. I Nervous system = central nervous system (CNS) + peripheral nervous system (PNS) Image: public domain (by Wikipedia User: Persion Poet Gal) Nervous System: CNS and PNS CNS PNS Chamberlain, Yannas, et al., 1998 Landstrom, Aria. “Nerve Regeneration Induced by Collagen-GAG Matrix in Collagen Tubes.” MS Thesis, MIT, 1994. Focus of interest: nerve fibers and axons Nerve fibers comprise axons wrapped in a myelin sheath, itself surrounded by BM (diam. 10-30 μm in rat sciatic nerve). Axons are extensions (long processes) of neurons located in spinal cord. They comprise endoplasmic reticulum and microtubules. 1. Anatomy and function of a peripheral nerve. II Myelinated axons (diam. 1-15 μm) are wrapped in a myelin sheath; nonmyelinated axons also exist. They are the elementary units for conduction of electric signals in the body. Myelin formed by wrapping a Schwann cell membrane many times around axon perimeter. No ECM inside nerve fibers. Myelin sheath is a wrapping of Schwann cell membranes around certain axons. 1. Anatomy and function of a peripheral nerve. III Nonmyelinated axons (diam. <1 μm) function in small pain nerves. Although surrounded by Schwann cells, they lack myelin sheath; Schwann cells are around them but have retained their cytoplasm. -

Guillain-Barre Syndrome (GBS)
Nervous tissue Anatomically Central nervous system (CNS) brain and spinal cord Peripheral nervous system (PNS) - cranial, spinal, and peripheral nerves - ganglia: nerve cell bodies outside the CNS Major cell types Neuron: nerve cell Supporting / Glial cells - Schwann cells, satellite cells (in PNS) - glia/neuroglia (in CNS) Neurone / Neuron Cell body Nucleus Cytoplasm (perikaryon) Process Axon Dendrites Axons (nerve fibers) Axon hillock Terminal boutons Dorsal root ganglia (DRG) nucleus ganglion/ganglia DRG neurons Basic neuron types Multipolar neuron Multiple dendrites Single axons Types: Interneurons Motor neurons Sympathetic neurons Bipolar neuron Single dendrite Single axon Types: Receptor neurons Vision Smell Balance Pseudo-unipolar neuron peripheral Single axon (Stem process) with stem 2 branches: Central process to spinal cord Peripheral process to terminal tissues (muscle, joints, skin et al) functionally: dendrite structurally: axon Type: Dorsal root ganglia (DRG neuron) central Pseudo-unipolar neuron Neuron: ultrastructure Rough endoplamic reticulum (rER) Nissl substance Cytoskeleton Microtubule Intermediate filaments: Neurofilaments Microfilaments: Actin Specialization of neuron/axon Cytoskeleton Axonal transport Neuron: ultrastructure rER: rough ER M: mitochondria L: lysosome G: Golgi Microscopic methods H & E (Hematoxylin and eosin) Nissl method Heavy metal impregnation Golgi, Cajal Thick sections / Spread preparations gold, silver: deposited in microtubules / neurofilaments Immunohistochemistry Microscopic methods: H & E -
Nervous System -I
Body Tissues Epithelial Connective Tissues Muscle Nervous Nervous system Controlling & Coordinating System Conducts nerve impulses between body structures and controls body functions Functions • Sensory Internal External • Integration> Analysis> storage>interpret>decide • Motor> Response • Regulates all activity (Voluntary & Involuntary) • Adjust according to changing external and internal environment Nervous System Subdivisions: CNS (Central Nervous System) PNS( Peripheral Nervous System) ANS (Autonomic Nervous system) Nervous tissue - Cell Types Functionally • Neuron (Nerve Cell) -Conduction Variable Shape , Size, Function • Neuroglia - Supportive -- Macroglia -- Microglia • Ependymal Cells • Schwann Cells - In PNS Neuron ( Nerve Cell) Components 1.Cell Body 2.Cell Processes (Neurites) Cell Body - Size vary from 5 µm - 120 µm (Perikaryon) – Plasma membrane Nucleus Cytoplasm Axon Hillock Neuronal Skeleton Cell Processes 1.Dendrites : Short , irregular thickness. Freely Branching, Afferent processes , Contain Nissl Granules 2. Axon – Long , Single, Efferent process of Uniform Diameter, Devoid of Nissl Granules, Ensheathed by Schwann cells, Gives collateral branches Terminal branches called telodendria (axon terminals) Terminate – within CNS - Always with another neuron Outside CNS – Either may end in relation to the effector organ or Synapse with neurons of Peripheral ganglia Types Of Neuron 1.Acc. To no of Processes Bipolar Multipolar Pseudounipolar 2. Acc. To Function Sensory Motor 3. Acc. To Axon Length Golgi type-1(long) Golgi type-II Synapse site of junction of neuron Types Axo- Dendritic Axo – Somatic Axo- Axonal Neuroglia • Astrocytes : Fibrous Protoplasmic Metabolism of neurotransmitters K+ Balance Contribute in brain development Blood brain barrier Link between neurons and blood vessels • Oligodendrocytes: Form a supporting network around neurons Produce myelin sheath around several neurons Neuroglia- contd. • Microglia: Phagocytic cells; Migrate to area of injured nervous tissue. -

The Spinal Cord and Spinal Nerves
14 The Nervous System: The Spinal Cord and Spinal Nerves PowerPoint® Lecture Presentations prepared by Steven Bassett Southeast Community College Lincoln, Nebraska © 2012 Pearson Education, Inc. Introduction • The Central Nervous System (CNS) consists of: • The spinal cord • Integrates and processes information • Can function with the brain • Can function independently of the brain • The brain • Integrates and processes information • Can function with the spinal cord • Can function independently of the spinal cord © 2012 Pearson Education, Inc. Gross Anatomy of the Spinal Cord • Features of the Spinal Cord • 45 cm in length • Passes through the foramen magnum • Extends from the brain to L1 • Consists of: • Cervical region • Thoracic region • Lumbar region • Sacral region • Coccygeal region © 2012 Pearson Education, Inc. Gross Anatomy of the Spinal Cord • Features of the Spinal Cord • Consists of (continued): • Cervical enlargement • Lumbosacral enlargement • Conus medullaris • Cauda equina • Filum terminale: becomes a component of the coccygeal ligament • Posterior and anterior median sulci © 2012 Pearson Education, Inc. Figure 14.1a Gross Anatomy of the Spinal Cord C1 C2 Cervical spinal C3 nerves C4 C5 C 6 Cervical C 7 enlargement C8 T1 T2 T3 T4 T5 T6 T7 Thoracic T8 spinal Posterior nerves T9 median sulcus T10 Lumbosacral T11 enlargement T12 L Conus 1 medullaris L2 Lumbar L3 Inferior spinal tip of nerves spinal cord L4 Cauda equina L5 S1 Sacral spinal S nerves 2 S3 S4 S5 Coccygeal Filum terminale nerve (Co1) (in coccygeal ligament) Superficial anatomy and orientation of the adult spinal cord. The numbers to the left identify the spinal nerves and indicate where the nerve roots leave the vertebral canal. -

A Review on Ultrasonic Neuromodulation of the Peripheral Nervous System: Enhanced Or Suppressed Activities?
applied sciences Review A Review on Ultrasonic Neuromodulation of the Peripheral Nervous System: Enhanced or Suppressed Activities? Bin Feng * , Longtu Chen and Sheikh J. Ilham Department of Biomedical Engineering, University of Connecticut, Storrs, CT 06269, USA; [email protected] (L.C.); [email protected] (S.J.I.) * Correspondence: [email protected]; Tel.: +1-860-486-6435 Received: 14 March 2019; Accepted: 9 April 2019; Published: 19 April 2019 Featured Application: The current review summarizes our recent knowledge of ultrasonic neuromodulation of peripheral nerve endings, axons, and somata in the dorsal root ganglion. This review indicates that focused ultrasound application at intermediate intensity can be a non-thermal and reversible neuromodulatory means for targeting the peripheral nervous system to manage neurological disorders. Abstract: Ultrasonic (US) neuromodulation has emerged as a promising therapeutic means by delivering focused energy deep into the nervous tissue. Low-intensity ultrasound (US) directly activates and/or inhibits neurons in the central nervous system (CNS). US neuromodulation of the peripheral nervous system (PNS) is less developed and rarely used clinically. The literature on the neuromodulatory effects of US on the PNS is controversial, with some studies documenting enhanced neural activities, some showing suppressed activities, and others reporting mixed effects. US, with different ranges of intensity and strength, is likely to generate distinct physical effects in the stimulated neuronal tissues, which underlies different experimental outcomes in the literature. In this review, we summarize all the major reports that document the effects of US on peripheral nerve endings, axons, and/or somata in the dorsal root ganglion. In particular, we thoroughly discuss the potential impacts of the following key parameters on the study outcomes of PNS neuromodulation by US: frequency, pulse repetition frequency, duty cycle, intensity, metrics for peripheral neural activities, and type of biological preparations used in the studies. -

Adaptive Self-Healing Electronic Epineurium for Chronic Bidirectional Neural Interfaces
ARTICLE https://doi.org/10.1038/s41467-020-18025-3 OPEN Adaptive self-healing electronic epineurium for chronic bidirectional neural interfaces Kang-Il Song 1,2,9, Hyunseon Seo1,3,9, Duhwan Seong4, Seunghoe Kim1, Ki Jun Yu5, Yu-Chan Kim1, ✉ ✉ ✉ Jinseok Kim1, Seok Joon Kwon6,7, Hyung-Seop Han1, Inchan Youn1,8 , Hyojin Lee1,8 & Donghee Son 4 Realizing a clinical-grade electronic medicine for peripheral nerve disorders is challenging owing to the lack of rational material design that mimics the dynamic mechanical nature 1234567890():,; of peripheral nerves. Electronic medicine should be soft and stretchable, to feasibly allow autonomous mechanical nerve adaptation. Herein, we report a new type of neural interface platform, an adaptive self-healing electronic epineurium (A-SEE), which can form compres- sive stress-free and strain-insensitive electronics-nerve interfaces and enable facile biofluid- resistant self-locking owing to dynamic stress relaxation and water-proof self-bonding properties of intrinsically stretchable and self-healable insulating/conducting materials, respectively. Specifically, the A-SEE does not need to be sutured or glued when implanted, thereby significantly reducing complexity and the operation time of microneurosurgery. In addition, the autonomous mechanical adaptability of the A-SEE to peripheral nerves can significantly reduce the mechanical mismatch at electronics-nerve interfaces, which minimizes nerve compression-induced immune responses and device failure. Though a small amount of Ag leaked from the A-SEE is observed in vivo (17.03 ppm after 32 weeks of implantation), we successfully achieved a bidirectional neural signal recording and stimulation in a rat sciatic nerve model for 14 weeks. -

Traumatic Injury to Peripheral Nerves
AAEM MINIMONOGRAPH 28 ABSTRACT: This article reviews the epidemiology and classification of traumatic peripheral nerve injuries, the effects of these injuries on nerve and muscle, and how electrodiagnosis is used to help classify the injury. Mecha- nisms of recovery are also reviewed. Motor and sensory nerve conduction studies, needle electromyography, and other electrophysiological methods are particularly useful for localizing peripheral nerve injuries, detecting and quantifying the degree of axon loss, and contributing toward treatment de- cisions as well as prognostication. © 2000 American Association of Electrodiagnostic Medicine. Published by John Wiley & Sons, Inc. Muscle Nerve 23: 863–873, 2000 TRAUMATIC INJURY TO PERIPHERAL NERVES LAWRENCE R. ROBINSON, MD Department of Rehabilitation Medicine, University of Washington, Seattle, Washington 98195 USA EPIDEMIOLOGY OF PERIPHERAL NERVE TRAUMA company central nervous system trauma, not only Traumatic injury to peripheral nerves results in con- compounding the disability, but making recognition siderable disability across the world. In peacetime, of the peripheral nerve lesion problematic. Of pa- peripheral nerve injuries commonly result from tients with peripheral nerve injuries, about 60% have 30 trauma due to motor vehicle accidents and less com- a traumatic brain injury. Conversely, of those with monly from penetrating trauma, falls, and industrial traumatic brain injury admitted to rehabilitation accidents. Of all patients admitted to Level I trauma units, 10 to 34% have associated peripheral nerve 7,14,39 centers, it is estimated that roughly 2 to 3% have injuries. It is often easy to miss peripheral nerve peripheral nerve injuries.30,36 If plexus and root in- injuries in the setting of central nervous system juries are also included, the incidence is about 5%.30 trauma. -

Introduction to the Peripheral Nervous System 2
Introduction to the 2 Peripheral Nervous System toward the neuron’s cell body, and an axon is the long process CONTENTS that carries the action potential away from the cell body. INTRODUCTION Some neurons appear to have only a single process extending PERIPHERAL NERVOUS SYSTEM from only one pole (a differentiated region of the cell body) SPINAL CORD (CENTRAL NERVOUS SYSTEM) that divides into two parts (Fig. 2-1). This type of neuron is OVERVIEW OF THE AUTONOMIC NERVOUS SYSTEM called a pseudounipolar neuron because embryonically it Sympathetic Nervous System develops from a bipolar neuroblast in which the two axons Parasympathetic Nervous System fuse. Multipolar neurons (Fig. 2-2) have multiple dendrites Visceral Afferent Neurons and typically a single axon arising from an enlarged portion REFERRED PAIN of the cell body called the axon hillock. These processes CLASSIFICATION OF NEURONAL FIBERS extend from different poles of the cell body. One neuron communicates with other neurons or glands DEVELOPMENT OF THE SPINAL CORD AND or muscle cells across a junction between cells called a PERIPHERAL NERVOUS SYSTEM synapse. Typically, communication is transmitted across a synapse by means of specific neurotransmitters, such as acetylcholine, epinephrine, and norepinephrine, but in some cases in the CNS by means of electric current passing ●●● INTRODUCTION from cell to cell. Many axons are ensheathed with a substance called The nervous system comprises the central nervous system myelin, which acts as an insulator. Myelinated axons transmit (CNS) and the peripheral nervous system (PNS). The CNS is impulses much faster than nonmyelinated axons. Myelin surrounded and protected by the skull (neurocranium) and consists of concentric layers of lipid-rich material formed vertebral column and consists of the brain and the spinal by the plasma membrane of a myelinating cell.