Neural Interfacing with Dorsal Root Ganglia: Anatomical Characterization and Electrophysiological Recordings with Novel Electrode Arrays
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Nerve Ultrasound in Dorsal Root Ganglion Disorders: Smaller Nerves Lead to Bigger Insights
Clinical Neurophysiology 130 (2019) 550–551 Contents lists available at ScienceDirect Clinical Neurophysiology journal homepage: www.elsevier.com/locate/clinph Editorial Nerve ultrasound in dorsal root ganglion disorders: Smaller nerves lead to bigger insights See Article, pages 568–572 After decades of having to make do with electric stimulation representing the fascicles, bundled together in a large outer cable and recording (i.e. nerve conduction studies, electromyography sheath (van Alfen et al., 2018). and evoked potentials), nerve ultrasound now provides the oppor- Next, it is important to realize what the ratio between axon/ tunity to improve neurodiagnostic patient care by deploying a myelin and connective tissue in a given nerve segment is, and powerful tool to detect neuromuscular pathology in an accurate how that ratio changes from the proximal root to the distal end and patient-friendly way (Mah et al., 2018; Walker et al., 2018). branches (Schraut et al., 2016). Connective tissue elements of the Nerve ultrasound is also increasingly providing neurologists and perineurium and epineurium are relatively sparse at the very prox- clinical neurophysiologists with the opportunity to increase their imal root and plexus levels, with an average connective tissue con- insight in the pathophysiology of peripheral nervous system tent of around 25–30%. Ultrasonographically, this means that roots (PNS) pathology. In this issue of Clinical Neurophysiology, Leadbet- will always look rather black in appearance without much dis- ter and coworkers (Leadbetter et al., 2019) describe the results of cernible fascicular architecture, as the sparseness of connective tis- their study on nerve ultrasound for diagnosing sensory neuronopa- sue elements provides relatively few reflectors to create an image thy in spinocerebellar ataxia type 2 and CANVAS syndrome. -
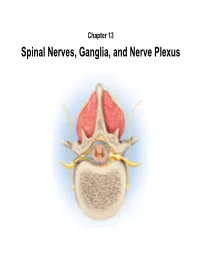
Spinal Nerves, Ganglia, and Nerve Plexus Spinal Nerves
Chapter 13 Spinal Nerves, Ganglia, and Nerve Plexus Spinal Nerves Posterior Spinous process of vertebra Posterior root Deep muscles of back Posterior ramus Spinal cord Transverse process of vertebra Posterior root ganglion Spinal nerve Anterior ramus Meningeal branch Communicating rami Anterior root Vertebral body Sympathetic ganglion Anterior General Anatomy of Nerves and Ganglia • Spinal cord communicates with the rest of the body by way of spinal nerves • nerve = a cordlike organ composed of numerous nerve fibers (axons) bound together by connective tissue – mixed nerves contain both afferent (sensory) and efferent (motor) fibers – composed of thousands of fibers carrying currents in opposite directions Anatomy of a Nerve Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Epineurium Perineurium Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Endoneurium Nerve Rootlets fiber Posterior root Fascicle Posterior root ganglion Anterior Blood root vessels Spinal nerve (b) Copyright by R.G. Kessel and R.H. Kardon, Tissues and Organs: A Text-Atlas of Scanning Electron Microscopy, 1979, W.H. Freeman, All rights reserved Blood vessels Fascicle Epineurium Perineurium Unmyelinated nerve fibers Myelinated nerve fibers (a) Endoneurium Myelin General Anatomy of Nerves and Ganglia • nerves of peripheral nervous system are ensheathed in Schwann cells – forms neurilemma and often a myelin sheath around the axon – external to neurilemma, each fiber is surrounded by -

The Peripheral Nervous System
The Peripheral Nervous System Dr. Ali Ebneshahidi Peripheral Nervous System (PNS) – Consists of 12 pairs of cranial nerves and 31 pairs of spinal nerves. – Serves as a critical link between the body and the central nervous system. – peripheral nerves contain an outermost layer of fibrous connective tissue called epineurium which surrounds a thinner layer of fibrous connective tissue called perineurium (surrounds the bundles of nerve or fascicles). Individual nerve fibers within the nerve are surrounded by loose connective tissue called endoneurium. Cranial Nerves Cranial nerves are direct extensions of the brain. Only Nerve I (olfactory) originates from the cerebrum, the remaining 11 pairs originate from the brain stem. Nerve I (Olfactory)- for the sense of smell (sensory). Nerve II (Optic)- for the sense of vision (sensory). Nerve III (Oculomotor)- for controlling muscles and accessory structures of the eyes ( primarily motor). Nerve IV (Trochlear)- for controlling muscles of the eyes (primarily motor). Nerve V (Trigeminal)- for controlling muscles of the eyes, upper and lower jaws and tear glands (mixed). Nerve VI (Abducens)- for controlling muscles that move the eye (primarily motor). Nerve VII (Facial) – for the sense of taste and controlling facial muscles, tear glands and salivary glands (mixed). Nerve VIII (Vestibulocochlear)- for the senses of hearing and equilibrium (sensory). Nerve IX (Glossopharyngeal)- for controlling muscles in the pharynx and to control salivary glands (mixed). Nerve X (Vagus)- for controlling muscles used in speech, swallowing, and the digestive tract, and controls cardiac and smooth muscles (mixed). Nerve XI (Accessory)- for controlling muscles of soft palate, pharynx and larynx (primarily motor). Nerve XII (Hypoglossal) for controlling muscles that move the tongue ( primarily motor). -
PN1 (Midha) Microanatomy of Peripheral Nerves-Part1.Pdf
Peripheral Nerve Basics • Consist of processes of cell bodies found in the DRG, anterior horn, and autonomic ganglia • Organized by several distinct connective tissue layers – the epineurium, perineurium, and endoneurium • Vascular supply provided by the vasa nervorum Peripheral Nerve Basics • Neuronal processes bound into fascicles by perineurium • Fascicles bound into nerves by epineurium • Endoneurium is a division of the perineurium which form thin layers of connective tissue surrounding neuronal fibers in a fascicle Sural nerve in cross-section Epineurium • Loose areolar tissue with sparse, longitudinally-oriented collagen fibers • Some elastic fibers where epineurium abuts perineurium • Able to accommodate a significant amount of nerve stretching and movement • Increases in thickness where nerves cross joints • Constitutes an increasing proportion of nerves as they increase in size • Epineurial fat helps cushion nerves from compressive injury • Decreased epineurial fat found in patients with diabetes Perineurium • Cellular component composed of laminated fibroblasts of up to 15 layers in thickness which are bounded by a basal lamina • Semi-permeable: inner lamellae have tight junctions, providing a barrier to intercellular transport of macromolecules – Tight junctions can be loosened with topical anaesthetics and with osmotic change Perineurium • Exhibits a slightly positive internal pressure – Fascicular contents herniate upon perineurial injury • Under tension longitudinally – Nerve segment shortens upon transection – may complicate surgical repair as nerve can be stretched only approximately 10% before being inhibited by collagen Endoneurium • Intrafascicular connective tissue consisting of a collagenous matrix in the interstitial space • Develops into partitions of dense connective tissue between diverging fascicles and eventually becomes perineurium when the fascicles separate • Collagen fibers are longitudinally-oriented and run along nerve fibers and capillaries. -

Canine Dorsal Root Ganglia Satellite Glial Cells Represent an Exceptional Cell Population with Astrocytic and Oligodendrocytic P
www.nature.com/scientificreports OPEN Canine dorsal root ganglia satellite glial cells represent an exceptional cell population with astrocytic and Received: 17 August 2017 Accepted: 6 October 2017 oligodendrocytic properties Published: xx xx xxxx W. Tongtako1,2, A. Lehmbecker1, Y. Wang1,2, K. Hahn1,2, W. Baumgärtner1,2 & I. Gerhauser 1 Dogs can be used as a translational animal model to close the gap between basic discoveries in rodents and clinical trials in humans. The present study compared the species-specifc properties of satellite glial cells (SGCs) of canine and murine dorsal root ganglia (DRG) in situ and in vitro using light microscopy, electron microscopy, and immunostainings. The in situ expression of CNPase, GFAP, and glutamine synthetase (GS) has also been investigated in simian SGCs. In situ, most canine SGCs (>80%) expressed the neural progenitor cell markers nestin and Sox2. CNPase and GFAP were found in most canine and simian but not murine SGCs. GS was detected in 94% of simian and 71% of murine SGCs, whereas only 44% of canine SGCs expressed GS. In vitro, most canine (>84%) and murine (>96%) SGCs expressed CNPase, whereas GFAP expression was diferentially afected by culture conditions and varied between 10% and 40%. However, GFAP expression was induced by bone morphogenetic protein 4 in SGCs of both species. Interestingly, canine SGCs also stimulated neurite formation of DRG neurons. These fndings indicate that SGCs represent an exceptional, intermediate glial cell population with phenotypical characteristics of oligodendrocytes and astrocytes and might possess intrinsic regenerative capabilities in vivo. Since the discovery of glial cells over a century ago, substantial progress has been made in understanding the origin, development, and function of the diferent types of glial cells in the central nervous system (CNS) and peripheral nervous system (PNS)1. -

Gamma Motor Neurons Survive and Exacerbate Alpha Motor Neuron Degeneration In
Gamma motor neurons survive and exacerbate alpha PNAS PLUS motor neuron degeneration in ALS Melanie Lalancette-Heberta,b, Aarti Sharmaa,b, Alexander K. Lyashchenkoa,b, and Neil A. Shneidera,b,1 aCenter for Motor Neuron Biology and Disease, Columbia University, New York, NY 10032; and bDepartment of Neurology, Columbia University, New York, NY 10032 Edited by Rob Brownstone, University College London, London, United Kingdom, and accepted by Editorial Board Member Fred H. Gage October 27, 2016 (received for review April 4, 2016) The molecular and cellular basis of selective motor neuron (MN) the muscle spindle and control the sensitivity of spindle afferent vulnerability in amyotrophic lateral sclerosis (ALS) is not known. In discharge (15); beta (β) skeletofusimotor neurons innervate both genetically distinct mouse models of familial ALS expressing intra- and extrafusal muscle (16). In addition to morphological mutant superoxide dismutase-1 (SOD1), TAR DNA-binding protein differences, distinct muscle targets, and the absence of primary 43 (TDP-43), and fused in sarcoma (FUS), we demonstrate selective afferent (IA)inputsonγ-MNs, these functional MN subtypes also degeneration of alpha MNs (α-MNs) and complete sparing of differ in their trophic requirements, and γ-MNs express high levels gamma MNs (γ-MNs), which selectively innervate muscle spindles. of the glial cell line-derived neurotropic factor (GDNF) receptor Resistant γ-MNs are distinct from vulnerable α-MNs in that they Gfrα1 (17). γ-MNs are also molecularly distinguished by the ex- lack synaptic contacts from primary afferent (IA) fibers. Elimination pression of other selective markers including the transcription α of these synapses protects -MNs in the SOD1 mutant, implicating factor Err3 (18), Wnt7A (19), the serotonin receptor 1d (5-ht1d) this excitatory input in MN degeneration. -

Dorsal Root Injury—A Model for Exploring Pathophysiology and Therapeutic Strategies in Spinal Cord Injury
cells Review Dorsal Root Injury—A Model for Exploring Pathophysiology and Therapeutic Strategies in Spinal Cord Injury Håkan Aldskogius * and Elena N. Kozlova Laboratory of Regenertive Neurobiology, Biomedical Center, Department of Neuroscience, Uppsala University, 75124 Uppsala, Sweden; [email protected] * Correspondence: [email protected] Abstract: Unraveling the cellular and molecular mechanisms of spinal cord injury is fundamental for our possibility to develop successful therapeutic approaches. These approaches need to address the issues of the emergence of a non-permissive environment for axonal growth in the spinal cord, in combination with a failure of injured neurons to mount an effective regeneration program. Experimental in vivo models are of critical importance for exploring the potential clinical relevance of mechanistic findings and therapeutic innovations. However, the highly complex organization of the spinal cord, comprising multiple types of neurons, which form local neural networks, as well as short and long-ranging ascending or descending pathways, complicates detailed dissection of mechanistic processes, as well as identification/verification of therapeutic targets. Inducing different types of dorsal root injury at specific proximo-distal locations provide opportunities to distinguish key components underlying spinal cord regeneration failure. Crushing or cutting the dorsal root allows detailed analysis of the regeneration program of the sensory neurons, as well as of the glial response at the dorsal root-spinal cord interface without direct trauma to the spinal cord. At the same time, a lesion at this interface creates a localized injury of the spinal cord itself, but with an initial Citation: Aldskogius, H.; Kozlova, neuronal injury affecting only the axons of dorsal root ganglion neurons, and still a glial cell response E.N. -

Lecture 19: Nerve Synthesis in Vivo (Regeneration)
Nerve synthesis in vivo (regeneration)* 1. Anatomy and function of a peripheral nerve. 2. Experimental parameters for study of induced regeneration. 3. Synthesis of myelinated axons and BM (nerve fibers) 4. Evidence (?) of synthesis of an endoneurium. 5. Synthesis of a nerve trunk (including summary of kinetics of synthesis). 6. Comparative regenerative activity of various reactants. _______ *Tissue and Organ Regeneration in Adults, Yannas, Springer, 2001, Ch. 6. 1. Anatomy and function of a peripheral nerve. I Nervous system = central nervous system (CNS) + peripheral nervous system (PNS) Image: public domain (by Wikipedia User: Persion Poet Gal) Nervous System: CNS and PNS CNS PNS Chamberlain, Yannas, et al., 1998 Landstrom, Aria. “Nerve Regeneration Induced by Collagen-GAG Matrix in Collagen Tubes.” MS Thesis, MIT, 1994. Focus of interest: nerve fibers and axons Nerve fibers comprise axons wrapped in a myelin sheath, itself surrounded by BM (diam. 10-30 μm in rat sciatic nerve). Axons are extensions (long processes) of neurons located in spinal cord. They comprise endoplasmic reticulum and microtubules. 1. Anatomy and function of a peripheral nerve. II Myelinated axons (diam. 1-15 μm) are wrapped in a myelin sheath; nonmyelinated axons also exist. They are the elementary units for conduction of electric signals in the body. Myelin formed by wrapping a Schwann cell membrane many times around axon perimeter. No ECM inside nerve fibers. Myelin sheath is a wrapping of Schwann cell membranes around certain axons. 1. Anatomy and function of a peripheral nerve. III Nonmyelinated axons (diam. <1 μm) function in small pain nerves. Although surrounded by Schwann cells, they lack myelin sheath; Schwann cells are around them but have retained their cytoplasm. -

Guillain-Barre Syndrome (GBS)
Nervous tissue Anatomically Central nervous system (CNS) brain and spinal cord Peripheral nervous system (PNS) - cranial, spinal, and peripheral nerves - ganglia: nerve cell bodies outside the CNS Major cell types Neuron: nerve cell Supporting / Glial cells - Schwann cells, satellite cells (in PNS) - glia/neuroglia (in CNS) Neurone / Neuron Cell body Nucleus Cytoplasm (perikaryon) Process Axon Dendrites Axons (nerve fibers) Axon hillock Terminal boutons Dorsal root ganglia (DRG) nucleus ganglion/ganglia DRG neurons Basic neuron types Multipolar neuron Multiple dendrites Single axons Types: Interneurons Motor neurons Sympathetic neurons Bipolar neuron Single dendrite Single axon Types: Receptor neurons Vision Smell Balance Pseudo-unipolar neuron peripheral Single axon (Stem process) with stem 2 branches: Central process to spinal cord Peripheral process to terminal tissues (muscle, joints, skin et al) functionally: dendrite structurally: axon Type: Dorsal root ganglia (DRG neuron) central Pseudo-unipolar neuron Neuron: ultrastructure Rough endoplamic reticulum (rER) Nissl substance Cytoskeleton Microtubule Intermediate filaments: Neurofilaments Microfilaments: Actin Specialization of neuron/axon Cytoskeleton Axonal transport Neuron: ultrastructure rER: rough ER M: mitochondria L: lysosome G: Golgi Microscopic methods H & E (Hematoxylin and eosin) Nissl method Heavy metal impregnation Golgi, Cajal Thick sections / Spread preparations gold, silver: deposited in microtubules / neurofilaments Immunohistochemistry Microscopic methods: H & E -

Human Sensory Neurons: Membrane Properties and Sensitization by Inflammatory Mediators
PAINÒ 155 (2014) 1861–1870 www.elsevier.com/locate/pain Human sensory neurons: Membrane properties and sensitization by inflammatory mediators Steve Davidson a,1, Bryan A. Copits a,1, Jingming Zhang b, Guy Page b, Andrea Ghetti b, ⇑ Robert W. Gereau IV a, a Washington University Pain Center and Department of Anesthesiology, Washington University School of Medicine, St Louis, MO 63110, USA b AnaBios Corporation, San Diego, CA 92109, USA Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article. article info abstract Article history: Biological differences in sensory processing between human and model organisms may present signifi- Received 25 April 2014 cant obstacles to translational approaches in treating chronic pain. To better understand the physiology Received in revised form 28 May 2014 of human sensory neurons, we performed whole-cell patch-clamp recordings from 141 human dorsal Accepted 20 June 2014 root ganglion (hDRG) neurons from 5 young adult donors without chronic pain. Nearly all small-diameter hDRG neurons (<50 lm) displayed an inflection on the descending slope of the action potential, a defining feature of rodent nociceptive neurons. A high proportion of hDRG neurons were responsive to the algo- Keywords: gens allyl isothiocyanate (AITC) and ATP, as well as the pruritogens histamine and chloroquine. We show Bradykinin that a subset of hDRG neurons responded to the inflammatory compounds bradykinin and prostaglandin Dorsal root ganglia Human E2 with action potential discharge and show evidence of sensitization including lower rheobase. Itch Compared to electrically evoked action potentials, chemically induced action potentials were triggered Nociception from less depolarized thresholds and showed distinct afterhyperpolarization kinetics. -
Nervous System -I
Body Tissues Epithelial Connective Tissues Muscle Nervous Nervous system Controlling & Coordinating System Conducts nerve impulses between body structures and controls body functions Functions • Sensory Internal External • Integration> Analysis> storage>interpret>decide • Motor> Response • Regulates all activity (Voluntary & Involuntary) • Adjust according to changing external and internal environment Nervous System Subdivisions: CNS (Central Nervous System) PNS( Peripheral Nervous System) ANS (Autonomic Nervous system) Nervous tissue - Cell Types Functionally • Neuron (Nerve Cell) -Conduction Variable Shape , Size, Function • Neuroglia - Supportive -- Macroglia -- Microglia • Ependymal Cells • Schwann Cells - In PNS Neuron ( Nerve Cell) Components 1.Cell Body 2.Cell Processes (Neurites) Cell Body - Size vary from 5 µm - 120 µm (Perikaryon) – Plasma membrane Nucleus Cytoplasm Axon Hillock Neuronal Skeleton Cell Processes 1.Dendrites : Short , irregular thickness. Freely Branching, Afferent processes , Contain Nissl Granules 2. Axon – Long , Single, Efferent process of Uniform Diameter, Devoid of Nissl Granules, Ensheathed by Schwann cells, Gives collateral branches Terminal branches called telodendria (axon terminals) Terminate – within CNS - Always with another neuron Outside CNS – Either may end in relation to the effector organ or Synapse with neurons of Peripheral ganglia Types Of Neuron 1.Acc. To no of Processes Bipolar Multipolar Pseudounipolar 2. Acc. To Function Sensory Motor 3. Acc. To Axon Length Golgi type-1(long) Golgi type-II Synapse site of junction of neuron Types Axo- Dendritic Axo – Somatic Axo- Axonal Neuroglia • Astrocytes : Fibrous Protoplasmic Metabolism of neurotransmitters K+ Balance Contribute in brain development Blood brain barrier Link between neurons and blood vessels • Oligodendrocytes: Form a supporting network around neurons Produce myelin sheath around several neurons Neuroglia- contd. • Microglia: Phagocytic cells; Migrate to area of injured nervous tissue. -

The Spinal Cord and Spinal Nerves
14 The Nervous System: The Spinal Cord and Spinal Nerves PowerPoint® Lecture Presentations prepared by Steven Bassett Southeast Community College Lincoln, Nebraska © 2012 Pearson Education, Inc. Introduction • The Central Nervous System (CNS) consists of: • The spinal cord • Integrates and processes information • Can function with the brain • Can function independently of the brain • The brain • Integrates and processes information • Can function with the spinal cord • Can function independently of the spinal cord © 2012 Pearson Education, Inc. Gross Anatomy of the Spinal Cord • Features of the Spinal Cord • 45 cm in length • Passes through the foramen magnum • Extends from the brain to L1 • Consists of: • Cervical region • Thoracic region • Lumbar region • Sacral region • Coccygeal region © 2012 Pearson Education, Inc. Gross Anatomy of the Spinal Cord • Features of the Spinal Cord • Consists of (continued): • Cervical enlargement • Lumbosacral enlargement • Conus medullaris • Cauda equina • Filum terminale: becomes a component of the coccygeal ligament • Posterior and anterior median sulci © 2012 Pearson Education, Inc. Figure 14.1a Gross Anatomy of the Spinal Cord C1 C2 Cervical spinal C3 nerves C4 C5 C 6 Cervical C 7 enlargement C8 T1 T2 T3 T4 T5 T6 T7 Thoracic T8 spinal Posterior nerves T9 median sulcus T10 Lumbosacral T11 enlargement T12 L Conus 1 medullaris L2 Lumbar L3 Inferior spinal tip of nerves spinal cord L4 Cauda equina L5 S1 Sacral spinal S nerves 2 S3 S4 S5 Coccygeal Filum terminale nerve (Co1) (in coccygeal ligament) Superficial anatomy and orientation of the adult spinal cord. The numbers to the left identify the spinal nerves and indicate where the nerve roots leave the vertebral canal.