Serendipita Restingae Sp. Nov. (Sebacinales): an Orchid Mycorrhizal Agaricomycete with Wide Host Range
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

<I>Serendipita Sacchari</I>
MYCOTAXON ISSN (print) 0093-4666 (online) 2154-8889 Mycotaxon, Ltd. ©2020 July–September 2020—Volume 135, pp. 579–587 https://doi.org/10.5248/135.579 Serendipita sacchari sp. nov. from a sugarcane rhizosphere in southern China Ling Xie1,2#, Yan-Yan Long1,2#, Yan Zhang1,2, Yan-Lu Chen1,2, Wen-Long Zhang2* 1Plant Protection Research Institute, Guangxi Academy of Agricultural Science, Nanning 530007, China 2 Microbiology Research Institute, Guangxi Academy of Agricultural Science, Nanning 530007, China *Correspondence to: [email protected] Abstract—We isolated a new species, proposed here as Serendipita sacchari, from a sugarcane rhizosphere in Guangxi Province, China. This species is characterized by its unstable nucleus numbers (1–15) in its chlamydospores versus their regular distribution in hyphal cells. ITS rDNA and combined LSU+ TEF1-α sequence analyses also support the uniqueness of this new plant symbiont. Key words—molecular phylogeny, Sebacinales, Serendipitaceae, taxonomy Introduction Serendipita P. Roberts (Basidiomycota, Sebacinales, Serendipitaceae), typified with S. vermifera (Oberw.) P. Roberts, originally comprised seven species (Roberts 1993). Two additional new species S. lyrica Trichiès (Trichiès 2003), and S. herbamans K. Riess & al. (Riess & al. 2014) have been proposed in this genus, and two anamorphic species in Piriformospora Sav. Verma & al. have been recombined as S. indica (Sav. Verma & al.) M. Weiss & al., and S. williamsii (Zuccaro & M. Weiss) M. Weiss & al. (Verma & al. 1998; Basiewicz & al. 2012; Weiß & al. 2016). Serendipita currently contains 11 species and DNA barcodes are widely accepted as an important tool in delineating species (Schoch & al. 2012; Riess & al. 2014). # Ling Xie & Yan-Yan Long contributed equally to this work. -

Morphology and Molecules: the Sebacinales, a Case Study
Mycol Progress DOI 10.1007/s11557-014-0983-1 ORIGINAL ARTICLE Morphology and molecules: the Sebacinales, a case study Franz Oberwinkler & Kai Riess & Robert Bauer & Sigisfredo Garnica Received: 4 April 2014 /Accepted: 8 April 2014 # German Mycological Society and Springer-Verlag Berlin Heidelberg 2014 Abstract Morphological and molecular discrepancies in the irregular germinating spores and inconspicuous cystidia, and biodiversity of monophyletic groups are challenging. The S. flagelliformis with flagelliform dikaryophyses from intention of this study was to find out whether the high S. epigaea s.str. Additional clades in Sebacina, based on molecular diversity in Sebacinales can be verified by micro- molecular differences, cannot be distinguished morphologi- morphological characteristics. Therefore, we carried out mo- cally at present. lecular and morphological studies on all generic type species of Sebacinales and additional representative taxa. Our results encouraged us to disentangle some phylogenetic and taxo- Introduction nomic discrepancies and to improve sebacinalean classifica- tions. This comprises generic circumscriptions and affilia- Based on longitudinally septate meiosporangia in their mature tions, as well as higher taxon groupings. At the family level, stage, sebacinoid fungi were originally grouped together with we redefined the Sebacinaceae, formerly the Sebacinales tremelloid and exidioid taxa. Sebacinales in the present cir- group A, and set it apart from the Sebacinales group B. For cumscription were reviewed in detail recently (Oberwinkler taxonomical purposes, it seems appropriate to refer et al. 2013). We refer to this publication for traditional classi- Paulisebacina, Craterocolla, Chaetospermum, fication of genera and interpretation of some species. Here, we Globulisebacina, Tremelloscypha, and Sebacina to the summarize data that accumulated within several years of Sebacinaceae and Piriformospora, and Serendipita to the intensive sampling, from morphological and molecular stud- Sebacinales group B. -

Non-Targeted Colonization by the Endomycorrhizal Fungus, Serendipita Vermifera, in Three Weeds Typically Co-Occurring with Switchgrass
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers from the Nebraska Center for Biotechnology Biotechnology, Center for 2018 Non-targeted Colonization by the Endomycorrhizal Fungus, Serendipita vermifera, in Three Weeds Typically Co-occurring with Switchgrass Prasun Ray Noble Research Institute, LLC Yingqing Guo Noble Research Institute, LLC Jaydeep Kolape Noble Research Institute, LLC Noble Research Institute, LLC, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/biotechpapers Part of the Biotechnology Commons, and the Molecular, Cellular, and Tissue Engineering Commons Ray, Prasun; Guo, Yingqing; Kolape, Jaydeep; and , "Non-targeted Colonization by the Endomycorrhizal Fungus, Serendipita vermifera, in Three Weeds Typically Co-occurring with Switchgrass" (2018). Papers from the Nebraska Center for Biotechnology. 24. https://digitalcommons.unl.edu/biotechpapers/24 This Article is brought to you for free and open access by the Biotechnology, Center for at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Papers from the Nebraska Center for Biotechnology by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. fpls-08-02236 January 6, 2018 Time: 16:25 # 1 ORIGINAL RESEARCH published: 09 January 2018 doi: 10.3389/fpls.2017.02236 Non-targeted Colonization by the Endomycorrhizal Fungus, Serendipita vermifera, in Three Weeds Typically Co-occurring with Switchgrass Prasun Ray, Yingqing Guo, Jaydeep Kolape and Kelly D. Craven* Noble Research Institute, LLC, Ardmore, OK, United States Serendipita vermifera (=Sebacina vermifera; isolate MAFF305830) is a mycorrhizal fungus originally isolated from the roots of an Australian orchid that we have previously shown to be beneficial in enhancing biomass yield and drought tolerance in switchgrass, an important bioenergy crop for cellulosic ethanol production in the United States. -

Pilzgattungen Europas
Pilzgattungen Europas - Liste 3: Notizbuchartige Auswahlliste zur Bestimmungsliteratur für Aphyllophorales und Heterobasidiomyceten (ohne cyphelloide Pilze und ohne Rost- und Brandpilze) Bernhard Oertel INRES Universität Bonn Auf dem Hügel 6 D-53121 Bonn E-mail: [email protected] 24.06.2011 Gattungen 1) Hauptliste 2) Liste der heute nicht mehr gebräuchlichen Gattungsnamen (Anhang) 1) Hauptliste Abortiporus Murr. 1904 (muss Loweomyces hier dazugeschlagen werden?): Lebensweise: Z.T. phytoparasitisch an Wurzeln von Bäumen Typus: A. distortus (Schw. : Fr.) Murr. [= Boletus distortus Schw. : Fr.; heute: A. biennis (Bull. : Fr.) Sing.; Anamorfe: Sporotrichopsis terrestris (Schulz.) Stalpers; Synonym der Anamorfe: Ceriomyces terrestris Schulz.] Bestimm. d. Gatt.: Bernicchia (2005), 68 u. 74 (auch Arten- Schlüssel); Bresinsky u. Besl (2003), 64; Hansen u. Knudsen 3 (1997), 220; Jülich (1984), 37-38 u. 328; Pegler (1973), The Fungi 4B, 404; Ryvarden u. Gilbertson (1993), Bd. 1, 70 u. 81 (auch Arten- Schlüssel) Abb.: 2) Lit.: Bollmann, Gminder u. Reil-CD (2007) Fidalgo, O. (1969), Revision ..., Rickia 4, 99-208 Jahn (1963), 65 Lohmeyer, T.R. (2000), Porlinge zwischen Inn und Salzach ..., Mycol. Bavarica 4, 33-47 Moser et al. (1985 ff.), Farbatlas (Gatt.-beschr.) Murrill (1904), Bull. Torrey Bot. Club 31, 421 Ryvarden u. Gilbertson (1993), Bd. 1, 81 s. ferner in 1) Abundisporus Ryv. 1999 [Europa?]: Typus: A. fuscopurpureus (Pers.) Ryv. (= Polyporus fuscopurpureus Pers.) Lit.: Ryvarden, L. ("1998", p. 1999), African polypores ..., Belg. J. Bot. 131 [Heinemann-Festschrift], 150- 155 (S. 154) s. ferner in 1) Acanthobasidium Oberw. 1965 (zu Aleurodiscus?): Typus: A. delicatum (Wakef.) Oberw. ex Jül. (= Aleurodiscus delicatus Wakef.) Bestimm. d. Gatt.: Bernicchia u. -

A Higher-Level Phylogenetic Classification of the Fungi
mycological research 111 (2007) 509–547 available at www.sciencedirect.com journal homepage: www.elsevier.com/locate/mycres A higher-level phylogenetic classification of the Fungi David S. HIBBETTa,*, Manfred BINDERa, Joseph F. BISCHOFFb, Meredith BLACKWELLc, Paul F. CANNONd, Ove E. ERIKSSONe, Sabine HUHNDORFf, Timothy JAMESg, Paul M. KIRKd, Robert LU¨ CKINGf, H. THORSTEN LUMBSCHf, Franc¸ois LUTZONIg, P. Brandon MATHENYa, David J. MCLAUGHLINh, Martha J. POWELLi, Scott REDHEAD j, Conrad L. SCHOCHk, Joseph W. SPATAFORAk, Joost A. STALPERSl, Rytas VILGALYSg, M. Catherine AIMEm, Andre´ APTROOTn, Robert BAUERo, Dominik BEGEROWp, Gerald L. BENNYq, Lisa A. CASTLEBURYm, Pedro W. CROUSl, Yu-Cheng DAIr, Walter GAMSl, David M. GEISERs, Gareth W. GRIFFITHt,Ce´cile GUEIDANg, David L. HAWKSWORTHu, Geir HESTMARKv, Kentaro HOSAKAw, Richard A. HUMBERx, Kevin D. HYDEy, Joseph E. IRONSIDEt, Urmas KO˜ LJALGz, Cletus P. KURTZMANaa, Karl-Henrik LARSSONab, Robert LICHTWARDTac, Joyce LONGCOREad, Jolanta MIA˛ DLIKOWSKAg, Andrew MILLERae, Jean-Marc MONCALVOaf, Sharon MOZLEY-STANDRIDGEag, Franz OBERWINKLERo, Erast PARMASTOah, Vale´rie REEBg, Jack D. ROGERSai, Claude ROUXaj, Leif RYVARDENak, Jose´ Paulo SAMPAIOal, Arthur SCHU¨ ßLERam, Junta SUGIYAMAan, R. Greg THORNao, Leif TIBELLap, Wendy A. UNTEREINERaq, Christopher WALKERar, Zheng WANGa, Alex WEIRas, Michael WEISSo, Merlin M. WHITEat, Katarina WINKAe, Yi-Jian YAOau, Ning ZHANGav aBiology Department, Clark University, Worcester, MA 01610, USA bNational Library of Medicine, National Center for Biotechnology Information, -

Sebacinales: a Hitherto Overlooked Cosm of Heterobasidiomycetes with a Broad Mycorrhizal Potential*
Mycol. Res. 108 (9): 1003–1010 (September 2004). f The British Mycological Society 1003 DOI: 10.1017/S0953756204000772 Printed in the United Kingdom. Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential* Michael WEISS1**, Marc-Andre´SELOSSE2, Karl-Heinz REXER3, Alexander URBAN4 and Franz OBERWINKLER1 1 Botanisches Institut, Universita¨tTu¨bingen, Auf der Morgenstelle 1, D-72076 Tu¨bingen, Germany. 2 UMR CNRS 7138, Syste´matique, Adaptation et Evolution, Muse´um d’Histoire Naturelle, 43 rue Cuvier, F-75005 Paris, France. 3 Fachbereich Biologie, Universita¨t Marburg, Karl-von Frisch-Straße 1, D-35032 Marburg, Germany. 4 Institut fu¨r Botanik, Universita¨t Wien, Rennweg 14, A-1030 Wien, Austria. E-mail : [email protected] Received 28 April 2004; accepted 16 June 2004. Within the basidiomycetes, the vast majority of known mycorrhizal species are homobasidiomycetes. It was therefore surprising when molecular and ultrastructural studies revealed a broad diversity of mycorrhizal associations involving members of the heterobasidiomycetous Sebacinaceae, fungi which, due to their inconspicuous basidiomes, have been often overlooked. To investigate the phylogenetic position of the Sebacinaceae within the basidiomycetes and to infer phylogenetic relationships within the Sebacinaceae, we made molecular phylogenetic analyses based on nuclear rDNA. We present a well-resolved phylogeny of the main lineages of basidiomycetes which suggests that the Sebacinaceae is the most basal group with known mycorrhizal members. Since more basal taxa of basidiomycetes consist of predominantly mycoparasitic and phytoparasitic fungi, it seems possible that a mycorrhizal life strategy, which was transformed into a saprotrophic strategy several times convergently, is an apomorphic character for the Hymenomycetidae. -

Notes, Outline and Divergence Times of Basidiomycota
Fungal Diversity (2019) 99:105–367 https://doi.org/10.1007/s13225-019-00435-4 (0123456789().,-volV)(0123456789().,- volV) Notes, outline and divergence times of Basidiomycota 1,2,3 1,4 3 5 5 Mao-Qiang He • Rui-Lin Zhao • Kevin D. Hyde • Dominik Begerow • Martin Kemler • 6 7 8,9 10 11 Andrey Yurkov • Eric H. C. McKenzie • Olivier Raspe´ • Makoto Kakishima • Santiago Sa´nchez-Ramı´rez • 12 13 14 15 16 Else C. Vellinga • Roy Halling • Viktor Papp • Ivan V. Zmitrovich • Bart Buyck • 8,9 3 17 18 1 Damien Ertz • Nalin N. Wijayawardene • Bao-Kai Cui • Nathan Schoutteten • Xin-Zhan Liu • 19 1 1,3 1 1 1 Tai-Hui Li • Yi-Jian Yao • Xin-Yu Zhu • An-Qi Liu • Guo-Jie Li • Ming-Zhe Zhang • 1 1 20 21,22 23 Zhi-Lin Ling • Bin Cao • Vladimı´r Antonı´n • Teun Boekhout • Bianca Denise Barbosa da Silva • 18 24 25 26 27 Eske De Crop • Cony Decock • Ba´lint Dima • Arun Kumar Dutta • Jack W. Fell • 28 29 30 31 Jo´ zsef Geml • Masoomeh Ghobad-Nejhad • Admir J. Giachini • Tatiana B. Gibertoni • 32 33,34 17 35 Sergio P. Gorjo´ n • Danny Haelewaters • Shuang-Hui He • Brendan P. Hodkinson • 36 37 38 39 40,41 Egon Horak • Tamotsu Hoshino • Alfredo Justo • Young Woon Lim • Nelson Menolli Jr. • 42 43,44 45 46 47 Armin Mesˇic´ • Jean-Marc Moncalvo • Gregory M. Mueller • La´szlo´ G. Nagy • R. Henrik Nilsson • 48 48 49 2 Machiel Noordeloos • Jorinde Nuytinck • Takamichi Orihara • Cheewangkoon Ratchadawan • 50,51 52 53 Mario Rajchenberg • Alexandre G. -

A Higher Level Classification of All Living Organisms
RESEARCH ARTICLE A Higher Level Classification of All Living Organisms Michael A. Ruggiero1*, Dennis P. Gordon2, Thomas M. Orrell1, Nicolas Bailly3, Thierry Bourgoin4, Richard C. Brusca5, Thomas Cavalier-Smith6, Michael D. Guiry7, Paul M. Kirk8 1 Integrated Taxonomic Information System, National Museum of Natural History, Smithsonian Institution, Washington, District of Columbia, United States of America, 2 National Institute of Water & Atmospheric Research, Wellington, New Zealand, 3 WorldFish—FIN, Los Baños, Philippines, 4 Institut Systématique, Evolution, Biodiversité (ISYEB), UMR 7205 MNHN-CNRS-UPMC-EPHE, Sorbonne Universités, Museum National d'Histoire Naturelle, 57, rue Cuvier, CP 50, F-75005, Paris, France, 5 Department of Ecology & Evolutionary Biology, University of Arizona, Tucson, Arizona, United States of America, 6 Department of Zoology, University of Oxford, Oxford, United Kingdom, 7 The AlgaeBase Foundation & Irish Seaweed Research Group, Ryan Institute, National University of Ireland, Galway, Ireland, 8 Mycology Section, Royal Botanic Gardens, Kew, London, United Kingdom * [email protected] Abstract We present a consensus classification of life to embrace the more than 1.6 million species already provided by more than 3,000 taxonomists’ expert opinions in a unified and coherent, OPEN ACCESS hierarchically ranked system known as the Catalogue of Life (CoL). The intent of this collab- orative effort is to provide a hierarchical classification serving not only the needs of the Citation: Ruggiero MA, Gordon DP, Orrell TM, Bailly CoL’s database providers but also the diverse public-domain user community, most of N, Bourgoin T, Brusca RC, et al. (2015) A Higher Level Classification of All Living Organisms. PLoS whom are familiar with the Linnaean conceptual system of ordering taxon relationships. -

Shift in Fungal Communities and Associated Enzyme Activities Along an Age Gradient of Managed Pinus Sylvestris Stands
The ISME Journal (2017) 11, 863–874 © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej ORIGINAL ARTICLE Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands Julia Kyaschenko1, Karina E Clemmensen2, Andreas Hagenbo2, Erik Karltun1 and Björn D Lindahl1 1Department of Soil and Environment, Swedish University of Agricultural Sciences, Uppsala, Sweden and 2Department of Forest Mycology and Plant Pathology, Uppsala BioCenter, Swedish University of Agricultural Sciences, Uppsala, Sweden Forestry reshapes ecosystems with respect to tree age structure, soil properties and vegetation composition. These changes are likely to be paralleled by shifts in microbial community composition with potential feedbacks on ecosystem functioning. Here, we assessed fungal communities across a chronosequence of managed Pinus sylvestris stands and investigated correlations between taxonomic composition and extracellular enzyme activities. Not surprisingly, clear-cutting had a negative effect on ectomycorrhizal fungal abundance and diversity. In contrast, clear-cutting favoured proliferation of saprotrophic fungi correlated with enzymes involved in holocellulose decomposition. During stand development, the re-establishing ectomycorrhizal fungal community shifted in composition from dominance by Atheliaceae in younger stands to Cortinarius and Russula species in older stands. Late successional ectomycorrhizal taxa correlated with enzymes involved -

Studies in the Stypella Vermiformis Group (Auriculariales, Basidiomycota)
Studies in the Stypella vermiformis group (Auriculariales, Basidiomycota) Viacheslav Spirin, Vera Malysheva, Danny Haelewaters & Karl-Henrik Larsson Antonie van Leeuwenhoek Journal of Microbiology ISSN 0003-6072 Volume 112 Number 5 Antonie van Leeuwenhoek (2019) 112:753-764 DOI 10.1007/s10482-018-01209-9 1 23 Your article is published under the Creative Commons Attribution license which allows users to read, copy, distribute and make derivative works, as long as the author of the original work is cited. You may self- archive this article on your own website, an institutional repository or funder’s repository and make it publicly available immediately. 1 23 Antonie van Leeuwenhoek (2019) 112:753–764 https://doi.org/10.1007/s10482-018-01209-9 (0123456789().,-volV)( 0123456789().,-volV) ORIGINAL PAPER Studies in the Stypella vermiformis group (Auriculariales, Basidiomycota) Viacheslav Spirin . Vera Malysheva . Danny Haelewaters . Karl-Henrik Larsson Received: 27 June 2018 / Accepted: 30 November 2018 / Published online: 8 December 2018 Ó The Author(s) 2018 Abstract Stypella vermiformis is a heterobasid- three species from two newly described genera. S. iomycete producing minute gelatinous basidiocarps vermiformis s.str. is distributed in temperate Europe on rotten wood of conifers in the Northern Hemi- and has small-sized basidia and basidiospores, and it is sphere. In the current literature, Stypella papillata, the placed in a new genus, Mycostilla. Another genus, genus type of Stypella (described from Brazil), is Stypellopsis, is created for two other species, the North treated as a taxonomic synonym of S. vermiformis.In American Stypellopsis farlowii, comb. nov., and the the present paper, we revise the type material of S. -

Divergence and Ranking of Taxa Across the Kingdoms Animalia, Fungi and Plantae
Mycosphere 7 (11): 1678–1689 (2016) www.mycosphere.org ISSN 2077 7019 Article – special issue Doi 10.5943/mycosphere/7/11/5 Copyright © Guizhou Academy of Agricultural Sciences Divergence and ranking of taxa across the kingdoms Animalia, Fungi and Plantae Samarakoon MC1,2,3, Hyde KD 1,3, Promputtha I2, Ariyawansa HA4, Hongsanan S1* 1Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai 57100, Thailand 2Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai 50200, Thailand 3Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, 132 Lanhei Road, Kunming 650201, China 4Guizhou Academy of Sciences, Guiyang, 550009, Guizhou Province, China Samarakoon MC, Hyde KD, Promputtha I, Ariyawansa HA, Hongsanan S. 2016 – Divergence and ranking of taxa across the kingdoms Animalia, Fungi and Plantae. Mycosphere 7(11), 1678–1689, Doi 10.5943/mycosphere/7/11/5 Abstract In science, species are grouped and ranked in kingdoms, phyla, classes, orders, families and genera and several other intermediate taxa, in a taxonomic hierarchy. However, the ranking of phyla, classes, orders and families across kingdoms is not linked and there is unlikely to be any correlation between these ranks in animals, fungi or plants. In a few recent studies, divergence times have been used to develop more natural groupings within ranks and it has been suggested that divergence times should be used as a universal criterion in high level ranking. It would therefore be desirable to develop more stable and standardized grouping of taxa in phyla, classes, orders and families across the kingdoms using divergence times. -
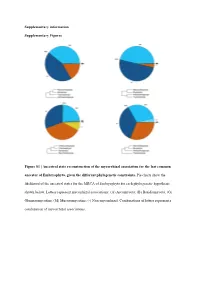
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea