A Comparison of Soils and Their Associated Microbial Communities
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Novel Bacterial Lineages Associated with Boreal Moss Species Hannah
bioRxiv preprint doi: https://doi.org/10.1101/219659; this version posted November 16, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Novel bacterial lineages associated with boreal moss species 2 Hannah Holland-Moritz1,2*, Julia Stuart3, Lily R. Lewis4, Samantha Miller3, Michelle C. Mack3, Stuart 3 F. McDaniel4, Noah Fierer1,2* 4 Affiliations: 5 1Cooperative Institute for Research in Environmental Sciences, University of Colorado at Boulder, 6 Boulder, CO, USA 7 2Department of Ecology and Evolutionary Biology, University of Colorado at Boulder, Boulder, CO, 8 USA 9 3Center for Ecosystem Science and Society, Northern Arizona University, Flagstaff, AZ USA 10 4Department of Biology, University of Florida, Gainesville, FL 32611-8525, USA 11 *Corresponding Author 12 13 Abstract 14 Mosses are critical components of boreal ecosystems where they typically account for a large 15 proportion of net primary productivity and harbor diverse bacterial communities that can be the major 16 source of biologically-fixed nitrogen in these ecosystems. Despite their ecological importance, we have 17 limited understanding of how microbial communities vary across boreal moss species and the extent to 18 which local environmental conditions may influence the composition of these bacterial communities. 19 We used marker gene sequencing to analyze bacterial communities associated with eight boreal moss 20 species collected near Fairbanks, AK USA. We found that host identity was more important than site in 21 determining bacterial community composition and that mosses harbor diverse lineages of potential N2- 22 fixers as well as an abundance of novel taxa assigned to understudied bacterial phyla (including 23 candidate phylum WPS-2). -

Genomic Analysis of Family UBA6911 (Group 18 Acidobacteria)
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.09.439258; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 2 Genomic analysis of family UBA6911 (Group 18 3 Acidobacteria) expands the metabolic capacities of the 4 phylum and highlights adaptations to terrestrial habitats. 5 6 Archana Yadav1, Jenna C. Borrelli1, Mostafa S. Elshahed1, and Noha H. Youssef1* 7 8 1Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, 9 OK 10 *Correspondence: Noha H. Youssef: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.04.09.439258; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 11 Abstract 12 Approaches for recovering and analyzing genomes belonging to novel, hitherto unexplored 13 bacterial lineages have provided invaluable insights into the metabolic capabilities and 14 ecological roles of yet-uncultured taxa. The phylum Acidobacteria is one of the most prevalent 15 and ecologically successful lineages on earth yet, currently, multiple lineages within this phylum 16 remain unexplored. Here, we utilize genomes recovered from Zodletone spring, an anaerobic 17 sulfide and sulfur-rich spring in southwestern Oklahoma, as well as from multiple disparate soil 18 and non-soil habitats, to examine the metabolic capabilities and ecological role of members of 19 the family UBA6911 (group18) Acidobacteria. -

Tree-Aggregated Predictive Modeling of Microbiome Data
www.nature.com/scientificreports OPEN Tree‑aggregated predictive modeling of microbiome data Jacob Bien1, Xiaohan Yan2, Léo Simpson3,4 & Christian L. Müller4,5,6* Modern high‑throughput sequencing technologies provide low‑cost microbiome survey data across all habitats of life at unprecedented scale. At the most granular level, the primary data consist of sparse counts of amplicon sequence variants or operational taxonomic units that are associated with taxonomic and phylogenetic group information. In this contribution, we leverage the hierarchical structure of amplicon data and propose a data‑driven and scalable tree‑guided aggregation framework to associate microbial subcompositions with response variables of interest. The excess number of zero or low count measurements at the read level forces traditional microbiome data analysis workfows to remove rare sequencing variants or group them by a fxed taxonomic rank, such as genus or phylum, or by phylogenetic similarity. By contrast, our framework, which we call trac (tree‑aggregation of compositional data), learns data‑adaptive taxon aggregation levels for predictive modeling, greatly reducing the need for user‑defned aggregation in preprocessing while simultaneously integrating seamlessly into the compositional data analysis framework. We illustrate the versatility of our framework in the context of large‑scale regression problems in human gut, soil, and marine microbial ecosystems. We posit that the inferred aggregation levels provide highly interpretable taxon groupings that can help microbiome researchers gain insights into the structure and functioning of the underlying ecosystem of interest. Microbial communities populate all major environments on earth and signifcantly contribute to the total plan- etary biomass. Current estimates suggest that a typical human-associated microbiome consists of ∼ 1013 bacteria1 and that marine bacteria and protists contribute to as much as 70% of the total marine biomass2. -

Microbial Communities of Polymetallic Deposits' Acidic
Microbial Communities of Polymetallic Deposits’ Acidic Ecosystems of ANGOR UNIVERSITY Continental Climatic Zone With High Temperature Contrasts Gavrilov, Sergei N.; Korzhenkov, Aleksei A.; Kublanov, Ilya V.; Bargiela, Rafael; Zamana, Leonid V.; Toshchakov, Stepan V.; Golyshin, Peter; Golyshina, Olga Frontiers in Microbiology PRIFYSGOL BANGOR / B Published: 17/07/2019 Publisher's PDF, also known as Version of record Cyswllt i'r cyhoeddiad / Link to publication Dyfyniad o'r fersiwn a gyhoeddwyd / Citation for published version (APA): Gavrilov, S. N., Korzhenkov, A. A., Kublanov, I. V., Bargiela, R., Zamana, L. V., Toshchakov, S. V., Golyshin, P., & Golyshina, O. (2019). Microbial Communities of Polymetallic Deposits’ Acidic Ecosystems of Continental Climatic Zone With High Temperature Contrasts. Frontiers in Microbiology. Hawliau Cyffredinol / General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. 01. Oct. 2021 ORIGINAL RESEARCH published: 17 July 2019 doi: 10.3389/fmicb.2019.01573 Microbial Communities of Polymetallic Deposits’ Acidic Ecosystems of Continental Climatic Zone With High Temperature Contrasts Sergey N. -
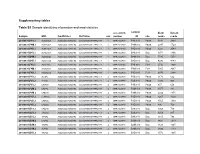
Supplementary Tables Table S1 Sample Identifying Information And
Supplementary tables Table S1 Sample identifying information and read statistics accession sample #raw #clean Sample MID FwdPrimer RvPrimer run number ID site reads reads 20100831.P4S.1 ACACGAGA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 P4S.910 Palsa 3831 2893 20100831.P4M.1 ACATAGCA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 P4M.910 Palsa 2047 1729 20100831.P4D.1 ACGATACA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 P4D.910 Palsa 3221 2543 20100831.B4S.1 ACTGCTCA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 B4S.910 Bog 1971 1366 20100831.B4M.1 AGCAGCGC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 B4M.910 Bog 5530 5251 20100831.B4D.1 AGCGTGCA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 B4D.910 Bog 4296 4143 20100831.F4S.1 AGTCTAGC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 F4S.910 Fen* 2716 1890 20100831.F4M.1 ATGACTGA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 F4M.910 Fen* 5005 3987 20100831.F4D.1 ATGCACGC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 F4D.910 Fen* 2870 2384 20100901.P1S.2 ACTGAT ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P1S.910 Palsa 1170 662 20100901.P2S.2 ATGTGT ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P2S.910 Palsa 1320 923 20100901.P3S.2 CACAGT ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P3S.910 Palsa 857 656 20100901.P2M.2 CACTAC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P2M.910 Palsa 1077 881 20100901.P3M.2 CAGCAT ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P3M.910 Palsa 2032 1474 20100901.P1D.2 CTAGAGC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT -

Pyrosequencing Investigation Into the Bacterial Community in Permafrost Soils Along the China-Russia Crude Oil Pipeline (CRCOP)
Pyrosequencing Investigation into the Bacterial Community in Permafrost Soils along the China-Russia Crude Oil Pipeline (CRCOP) Sizhong Yang1*, Xi Wen2, Huijun Jin1, Qingbai Wu1 1 State Key Laboratory of Frozen Soil Engineering (SKLFSE), Cold and Arid Regions Environmental and Engineering Research Institute (CAREERI), Chinese Academy of Sciences, Lanzhou, China, 2 College of Electrical Engineering, Northwest University for Nationalities, Lanzhou, China Abstract The China-Russia Crude Oil Pipeline (CRCOP) goes through 441 km permafrost soils in northeastern China. The bioremediation in case of oil spills is a major concern. So far, little is known about the indigenous bacteria inhabiting in the permafrost soils along the pipeline. A pilot 454 pyrosequencing analysis on the communities from four selected sites which possess high environment risk along the CRCOP is herein presented. The results reveal an immense bacterial diversity than previously anticipated. A total of 14448 OTUs with 84834 reads are identified, which could be assigned into 39 different phyla, and 223 families or 386 genera. Only five phyla sustain a mean OTU abundance more than 5% in all the samples, but they altogether account for 85.08% of total reads. Proteobacteria accounts for 41.65% of the total OTUs or 45% of the reads across all samples, and its proportion generally increases with soil depth, but OTUs numerically decline. Among Proteobacteria, the abundance of Beta-, Alpha-, Delta- and Gamma- subdivisions average to 38.7% (2331 OTUs), 37.5% (2257 OTUs), 10.35% (616 OTUs), and 6.21% (374 OTUs), respectively. Acidobacteria (esp. Acidobacteriaceae), Actinobacteria (esp. Intrasporangiaceae), Bacteroidetes (esp. Sphingobacteria and Flavobacteria) and Chloroflexi (esp. -

The Effect of Tillage System and Crop Rotation on Soil Microbial Diversity and Composition in a Subtropical Acrisol
Diversity 2012, 4, 375-395; doi:10.3390/d4040375 OPEN ACCESS diversity ISSN 1424-2818 www.mdpi.com/journal/diversity Article The Effect of Tillage System and Crop Rotation on Soil Microbial Diversity and Composition in a Subtropical Acrisol Patricia Dorr de Quadros 1,2,*, Kateryna Zhalnina 2, Austin Davis-Richardson 2, Jennie R. Fagen 2, Jennifer Drew 2, Cimelio Bayer 1, Flavio A.O. Camargo 1 and Eric W. Triplett 2 1 Department of Soil Science, Federal University of Rio Grande do Sul, Porto Alegre, Brazil; E-Mails: [email protected] (C.B.); [email protected] (F.A.O.C.) 2 Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences, University of Florida, Gainesville, FL, USA; E-Mails: [email protected] (K.Z.), [email protected] (A.C.R.), [email protected] (J.D.), [email protected] (E.W.T.) *Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: (352)-575-5731. Received: 29 August 2012; in revised form: 11 October 2012 / Accepted: 19 October 2012 / Published: 31 October 2012 Abstract: Agricultural management alters physical and chemical soil properties, which directly affects microbial life strategies and community composition. The microbial community drives important nutrient cycling processes that can influence soil quality, cropping productivity and environmental sustainability. In this research, a long-term agricultural experiment in a subtropical Acrisol was studied in south Brazil. The plots at this site represent two tillage systems, two nitrogen fertilization regimes and three crop rotation systems. Using Illumina high-throughput sequencing of the 16S rRNA gene, the archaeal and bacterial composition was determined from phylum to species level in the different plot treatments. -

The First Representative of the Globally Widespread Subdivision 6 Acidobacteria, Vicinamibacter Silvestris Gen
International Journal of Systematic and Evolutionary Microbiology (2016), 66, 2971–2979 DOI 10.1099/ijsem.0.001131 The first representative of the globally widespread subdivision 6 Acidobacteria, Vicinamibacter silvestris gen. nov., sp. nov., isolated from subtropical savannah soil Katharina J. Huber,1 Alicia M. Geppert,1 Gerhard Wanner,2 Barbel€ U. Fösel,1 Pia K. Wüst1 and Jörg Overmann1,3 Correspondence 1Department of Microbial Ecology and Diversity Research, Leibniz Institute DSMZ – German Katharina J. Huber Collection of Microorganisms and Cell Cultures, Braunschweig, Germany [email protected] 2Department of Biology I, Biozentrum Ludwig Maximilian University of Munich, Planegg- Martinsried, Germany 3Technical University Braunschweig, Braunschweig, Germany Members of the phylum Acidobacteria are abundant in a wide variety of soil environments. Despite this, previous cultivation attempts have frequently failed to retrieve representative phylotypes of Acidobacteria, which have, therefore, been discovered by culture-independent methods (13175 acidobacterial sequences in the SILVA database version 123; NR99) and only 47 species have been described so far. Strain Ac_5_C6T represents the first isolate of the globally widespread and abundant subdivision 6 Acidobacteria and is described in the present study. Cells of strain Ac_5_C6T were Gram-stain-negative, immotile rods that divided by binary fission. They formed yellow, extremely cohesive colonies and stable aggregates even in rapidly shaken liquid cultures. Ac_5_C6T was tolerant of a wide range of temperatures (12–40 C) and pH values (4.7–9.0). It grew chemoorganoheterotrophically on a broad range of substrates including different sugars, organic acids, nucleic acids and complex proteinaceous compounds. T The major fatty acids of Ac_5_C6 were iso-C17 : 1 !9c,C18 : 1 !7c and iso-C15 : 0. -

In Danish Wastewater Treatment Plants
fmicb-12-643950 April 16, 2021 Time: 17:35 # 1 ORIGINAL RESEARCH published: 22 April 2021 doi: 10.3389/fmicb.2021.643950 High Diversity and Functional Potential of Undescribed “Acidobacteriota” in Danish Wastewater Treatment Plants Jannie Munk Kristensen†, Caitlin Singleton, Lee-Ann Clegg, Francesca Petriglieri and Per Halkjaer Nielsen* Department of Chemistry and Bioscience, Center for Microbial Communities, Aalborg University, Aalborg, Denmark Microbial communities in water resource recovery facilities encompass a large diversity Edited by: of poorly characterized lineages that could have undescribed process-critical functions. Bärbel Ulrike Fösel, Recently, it was shown that taxa belonging to “Acidobacteriota” are abundant in Helmholtz Zentrum München, Helmholtz-Gemeinschaft Deutscher Danish full-scale activated sludge wastewater treatment plants (WWTP), and here we Forschungszentren, Germany investigated their diversity, distribution, and functional potential. “Acidobacteriota” taxa Reviewed by: were identified using a comprehensive full-length 16S rRNA gene reference dataset Aharon Oren, and amplicon sequencing surveys across 37 WWTPs. Members of this phylum were Hebrew University of Jerusalem, Israel Andreas Schramm, diverse, belonging to 14 families, eight of which are completely uncharacterized and Aarhus University, Denmark lack type strains. Several lineages were abundant, with relative abundances of up to *Correspondence: 5% of the microbial community. Genome annotation and metabolic reconstruction of Per Halkjaer Nielsen [email protected] 50 high-quality “Acidobacteriota” metagenome-assembled genomes (MAGs) from 19 †Present address: WWTPs showed high metabolic diversity and potential involvement in nitrogen and Jannie Munk Kristensen, phosphorus removal and iron reduction. Fluorescence in situ hybridization (FISH) using Division of Microbial Ecology, Center newly-designed probes revealed cells with diverse morphologies, predominantly located for Microbiology and Environmental Systems Science, University inside activated sludge flocs. -

Microbial Community Structure Analysis in Acer Palmatum Bark and Isolation of Novel Bacteria IAD-21 of the Phylum Abditibacteriota (Former Candidate Division FBP)
Microbial community structure analysis in Acer palmatum bark and isolation of novel bacteria IAD-21 of the phylum Abditibacteriota (former candidate division FBP) Kazuki Kobayashi1 and Hideki Aoyagi1,2 1 Division of Life Sciences and Bioengineering, Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Ibaraki, Japan 2 Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Ibaraki, Japan ABSTRACT Background: The potential of unidentified microorganisms for academic and other applications is limitless. Plants have diverse microbial communities associated with their biomes. However, few studies have focused on the microbial community structure relevant to tree bark. Methods: In this report, the microbial community structure of bark from the broad-leaved tree Acer palmatum was analyzed. Both a culture-independent approach using polymerase chain reaction (PCR) amplification and next generation sequencing, and bacterial isolation and sequence-based identification methods were used to explore the bark sample as a source of previously uncultured microorganisms. Molecular phylogenetic analyses based on PCR-amplified 16S rDNA sequences were performed. Results: At the phylum level, Proteobacteria and Bacteroidetes were relatively abundant in the A. palmatum bark. In addition, microorganisms from the phyla Acidobacteria, Gemmatimonadetes, Verrucomicrobia, Armatimonadetes, and Submitted 2 February 2019 Abditibacteriota, which contain many uncultured microbial species, existed in the Accepted 12 September 2019 A. palmatum bark. Of the 30 genera present at relatively high abundance in the bark, Published 29 October 2019 some genera belonging to the phyla mentioned were detected. A total of 70 isolates Corresponding author could be isolated and cultured using the low-nutrient agar media DR2A and Hideki Aoyagi, PE03. -
Land Use Effects on Airborne Bacterial Communities Are Evident in Both Near-Surface and Higher-Altitude Air
diversity Article Land Use Effects on Airborne Bacterial Communities Are Evident in Both Near-Surface and Higher-Altitude Air Allison M. Spring 1, Kenneth D. Domingue 2 , Thomas V. Kerber 2, Margaret M. Mooney 2, Rebecca L. Hale 3, Kristina M. Lemmer 2 and Kathryn M. Docherty 1,* 1 Department of Biological Sciences, Western Michigan University, Kalamazoo, MI 49006, USA; [email protected] 2 Department of Mechanical and Aerospace Engineering, Western Michigan University, Kalamazoo, MI 49006, USA; [email protected] (K.D.D.); [email protected] (T.V.K.); [email protected] (M.M.M.); [email protected] (K.M.L.) 3 Department of Biological Sciences, Idaho State University, Pocatello, ID 83209, USA; [email protected] * Correspondence: [email protected] Abstract: Land use influences the composition of near-surface airborne bacterial communities, and bacteria can be transported through the atmosphere at global scales. The atmosphere mixes vertically, but rigorously assessing whether the effects of land use on atmospheric communities extends to higher altitudes requires examining communities from multiple altitudes collected at a stable location and timeframe. In this study, we collected near-surface (<2 m) and higher-altitude (150 m) air samples from three sites in an agricultural/developed location and a forested/undeveloped location. We used bacterial 16S rRNA amplicon sequencing to compare communities and predict functionality by altitude. Higher-altitude and near-surface communities did not differ in composition within each Citation: Spring, A.M.; Domingue, location. Communities collected above the undeveloped location were equally variable at both K.D.; Kerber, T.V.; Mooney, M.M.; altitudes; higher-altitude samples from the developed location predominantly contained Firmicutes Hale, R.L.; Lemmer, K.M.; Docherty, K.M. -
The Microbial Ecology and Horticultural Sustainability of Organically and Conventionally Managed Apples
ABSTRACT Title of Document: THE MICROBIAL ECOLOGY AND HORTICULTURAL SUSTAINABILITY OF ORGANICALLY AND CONVENTIONALLY MANAGED APPLES. Andrea R. Ottesen, PhD, 2008 Directed By: Professor, Dr. Christopher S. Walsh, Plant Sciences and Landscape Architecture Objectives Organically and conventionally managed apple trees (Malus domestica Borkh) were evaluated for three growing seasons (2005-2007) to examine the impact of organic and conventional pesticide applications on the microbial ecology of phyllosphere and soil microflora. An important objective was to establish if organic or conventional selection pressures contribute to an increased presence of enteric pathogens in phyllosphere microflora. The horticultural and economic sustainability of the organic crop was also compared to the conventional crop with regard to fruit yield and input costs. Methods Microbial populations from phyllosphere and soil environments of apple trees were evaluated using clone libraries of 16S rRNA gene fragments. Clones were sequenced and software was used to assess diversity indices, identify shared similarities and compute statistical differences between communities. These measurements were subsequently used to examine treatment effects on the microbial libraries. Phyllosphere Results Eight bacterial phyla and 14 classes were found in this environment. A statistically significant difference between organically and conventionally managed phyllosphere bacterial microbial communities was observed at four of six sampling time points. Unique phylotypes were found associated with each management treatment but no increased human health risk could be associated with either treatment with regard to enteric pathogens. Soil Results Seventeen bacterial phyla spanning twenty-two classes, and two archaeal phyla spanning eight classes, were seen in the 16S rRNA gene libraries of organic and conventional soil samples.