Biodegradation Potential and Diversity of Diclofenac-Degrading Microbiota in an Immobilized Cell Biofilter
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Lichens' Microbiota, Still a Mystery?
fmicb-12-623839 March 24, 2021 Time: 15:25 # 1 REVIEW published: 30 March 2021 doi: 10.3389/fmicb.2021.623839 The Lichens’ Microbiota, Still a Mystery? Maria Grimm1*, Martin Grube2, Ulf Schiefelbein3, Daniela Zühlke1, Jörg Bernhardt1 and Katharina Riedel1 1 Institute of Microbiology, University Greifswald, Greifswald, Germany, 2 Institute of Plant Sciences, Karl-Franzens-University Graz, Graz, Austria, 3 Botanical Garden, University of Rostock, Rostock, Germany Lichens represent self-supporting symbioses, which occur in a wide range of terrestrial habitats and which contribute significantly to mineral cycling and energy flow at a global scale. Lichens usually grow much slower than higher plants. Nevertheless, lichens can contribute substantially to biomass production. This review focuses on the lichen symbiosis in general and especially on the model species Lobaria pulmonaria L. Hoffm., which is a large foliose lichen that occurs worldwide on tree trunks in undisturbed forests with long ecological continuity. In comparison to many other lichens, L. pulmonaria is less tolerant to desiccation and highly sensitive to air pollution. The name- giving mycobiont (belonging to the Ascomycota), provides a protective layer covering a layer of the green-algal photobiont (Dictyochloropsis reticulata) and interspersed cyanobacterial cell clusters (Nostoc spec.). Recently performed metaproteome analyses Edited by: confirm the partition of functions in lichen partnerships. The ample functional diversity Nathalie Connil, Université de Rouen, France of the mycobiont contrasts the predominant function of the photobiont in production Reviewed by: (and secretion) of energy-rich carbohydrates, and the cyanobiont’s contribution by Dirk Benndorf, nitrogen fixation. In addition, high throughput and state-of-the-art metagenomics and Otto von Guericke University community fingerprinting, metatranscriptomics, and MS-based metaproteomics identify Magdeburg, Germany Guilherme Lanzi Sassaki, the bacterial community present on L. -

Novel Bacterial Lineages Associated with Boreal Moss Species Hannah
bioRxiv preprint doi: https://doi.org/10.1101/219659; this version posted November 16, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Novel bacterial lineages associated with boreal moss species 2 Hannah Holland-Moritz1,2*, Julia Stuart3, Lily R. Lewis4, Samantha Miller3, Michelle C. Mack3, Stuart 3 F. McDaniel4, Noah Fierer1,2* 4 Affiliations: 5 1Cooperative Institute for Research in Environmental Sciences, University of Colorado at Boulder, 6 Boulder, CO, USA 7 2Department of Ecology and Evolutionary Biology, University of Colorado at Boulder, Boulder, CO, 8 USA 9 3Center for Ecosystem Science and Society, Northern Arizona University, Flagstaff, AZ USA 10 4Department of Biology, University of Florida, Gainesville, FL 32611-8525, USA 11 *Corresponding Author 12 13 Abstract 14 Mosses are critical components of boreal ecosystems where they typically account for a large 15 proportion of net primary productivity and harbor diverse bacterial communities that can be the major 16 source of biologically-fixed nitrogen in these ecosystems. Despite their ecological importance, we have 17 limited understanding of how microbial communities vary across boreal moss species and the extent to 18 which local environmental conditions may influence the composition of these bacterial communities. 19 We used marker gene sequencing to analyze bacterial communities associated with eight boreal moss 20 species collected near Fairbanks, AK USA. We found that host identity was more important than site in 21 determining bacterial community composition and that mosses harbor diverse lineages of potential N2- 22 fixers as well as an abundance of novel taxa assigned to understudied bacterial phyla (including 23 candidate phylum WPS-2). -

Edaphobacter Dinghuensis Sp. Nov., an Acidobacterium Isolated from Lower Subtropical Forest Soil Jia Wang,3 Mei-Hong Chen,3 Ying-Ying Lv, Ya-Wen Jiang and Li-Hong Qiu
International Journal of Systematic and Evolutionary Microbiology (2016), 66, 276–282 DOI 10.1099/ijsem.0.000710 Edaphobacter dinghuensis sp. nov., an acidobacterium isolated from lower subtropical forest soil Jia Wang,3 Mei-hong Chen,3 Ying-ying Lv, Ya-wen Jiang and Li-hong Qiu Correspondence State Key Laboratory of Biocontrol, School of Life Sciences, Sun Yat-sen University, Li-hong Qiu Guangzhou 510275, PR China [email protected] An aerobic bacterium, designated DHF9T, was isolated from a soil sample collected from the lower subtropical forest of Dinghushan Biosphere Reserve, Guangdong Province, PR China. Cells were Gram-stain-negative, non-motile, short rods that multiplied by binary division. Strain DHF9T was an obligately acidophilic, mesophilic bacterium capable of growth at pH 3.5–5.5 (optimum pH 4.0) and at 10–33 8C (optimum 28–33 8C). Growth was inhibited at NaCl concentrations above 2.0 % (w/v). The major fatty acids were iso-C15 : 0,C16 : 0 and C16 : 1v7c. The polar lipids consist of phosphatidylethanolamine, two unidentified aminolipids, two unidentified phospholipids, two unidentified polar lipids and an unidentified glycolipid. The DNA G+C content was 57.7 mol%. Phylogenetic analysis based on 16S rRNA gene sequences indicated that the strain belongs to the genus Edaphobacter in subdivision 1 of the phylum Acidobacteria, with the highest 16S rRNA gene sequence similarity of 97.0 % to Edaphobacter modestus Jbg-1T. Based on phylogenetic, chemotaxonomic and physiological analyses, it is proposed that strain DHF9T represents a novel species of the genus Edaphobacter, named Edaphobacter dinghuensis sp. nov. -

Genomic Analysis of Family UBA6911 (Group 18 Acidobacteria)
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.09.439258; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 2 Genomic analysis of family UBA6911 (Group 18 3 Acidobacteria) expands the metabolic capacities of the 4 phylum and highlights adaptations to terrestrial habitats. 5 6 Archana Yadav1, Jenna C. Borrelli1, Mostafa S. Elshahed1, and Noha H. Youssef1* 7 8 1Department of Microbiology and Molecular Genetics, Oklahoma State University, Stillwater, 9 OK 10 *Correspondence: Noha H. Youssef: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.04.09.439258; this version posted April 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 11 Abstract 12 Approaches for recovering and analyzing genomes belonging to novel, hitherto unexplored 13 bacterial lineages have provided invaluable insights into the metabolic capabilities and 14 ecological roles of yet-uncultured taxa. The phylum Acidobacteria is one of the most prevalent 15 and ecologically successful lineages on earth yet, currently, multiple lineages within this phylum 16 remain unexplored. Here, we utilize genomes recovered from Zodletone spring, an anaerobic 17 sulfide and sulfur-rich spring in southwestern Oklahoma, as well as from multiple disparate soil 18 and non-soil habitats, to examine the metabolic capabilities and ecological role of members of 19 the family UBA6911 (group18) Acidobacteria. -

Diversity of Biodeteriorative Bacterial and Fungal Consortia in Winter and Summer on Historical Sandstone of the Northern Pergol
applied sciences Article Diversity of Biodeteriorative Bacterial and Fungal Consortia in Winter and Summer on Historical Sandstone of the Northern Pergola, Museum of King John III’s Palace at Wilanow, Poland Magdalena Dyda 1,2,* , Agnieszka Laudy 3, Przemyslaw Decewicz 4 , Krzysztof Romaniuk 4, Martyna Ciezkowska 4, Anna Szajewska 5 , Danuta Solecka 6, Lukasz Dziewit 4 , Lukasz Drewniak 4 and Aleksandra Skłodowska 1 1 Department of Geomicrobiology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Miecznikowa 1, 02-096 Warsaw, Poland; [email protected] 2 Research and Development for Life Sciences Ltd. (RDLS Ltd.), Miecznikowa 1/5a, 02-096 Warsaw, Poland 3 Laboratory of Environmental Analysis, Museum of King John III’s Palace at Wilanow, Stanislawa Kostki Potockiego 10/16, 02-958 Warsaw, Poland; [email protected] 4 Department of Environmental Microbiology and Biotechnology, Institute of Microbiology, Faculty of Biology, University of Warsaw, Miecznikowa 1, 02-096 Warsaw, Poland; [email protected] (P.D.); [email protected] (K.R.); [email protected] (M.C.); [email protected] (L.D.); [email protected] (L.D.) 5 The Main School of Fire Service, Slowackiego 52/54, 01-629 Warsaw, Poland; [email protected] 6 Department of Plant Molecular Ecophysiology, Institute of Experimental Plant Biology and Biotechnology, Faculty of Biology, University of Warsaw, Miecznikowa 1, 02-096 Warsaw, Poland; [email protected] * Correspondence: [email protected] or [email protected]; Tel.: +48-786-28-44-96 Citation: Dyda, M.; Laudy, A.; Abstract: The aim of the presented investigation was to describe seasonal changes of microbial com- Decewicz, P.; Romaniuk, K.; munity composition in situ in different biocenoses on historical sandstone of the Northern Pergola in Ciezkowska, M.; Szajewska, A.; the Museum of King John III’s Palace at Wilanow (Poland). -

Biodegradation of Fluorinated Compounds Widely Used in Agro-Industrial Applications Diogo Alves Da Mota Alexandrino M 2016
DISSERTAÇÃO DE MESTRADO TOXICOLOGIA E CONTAMINAÇÃO AMBIENTAIS Biodegradation of fluorinated compounds widely used in agro-industrial applications Diogo Alves da Mota Alexandrino M 2016 Diogo Alves da Mota Alexandrino BIODEGRADATION OF FLUORINATED COMPOUNDS WIDELY USED IN AGRO-INDUSTRIAL CONTEXTS Dissertação de Candidatura ao grau de Mestre em Toxicologia e Contaminação Ambientais submetida ao Instituto de Ciências Biomédicas de Abel Salazar da Universidade do Porto. Orientadora – Doutora Maria de Fátima Carvalho Categoria – Investigadora Auxiliar Afiliação – Centro Interdisciplinar de Investigação Marinha e Ambiental da Universidade do Porto Co-orientadora – Doutora Ana Paula Mucha Categoria – Investigadora Auxiliar Afiliação – Centro Interdisciplinar de Investigação Marinha e Ambiental da Universidade do Porto ACKNOWLEDGEMENTS Firstly, I would like to thank my supervisor, Dr. Maria F. Carvalho, without whom the work integrated in this thesis would have not been possible. I genuinely thank her incredible dedication and trust, certain that part of my future goals have been established as a result of her mentoring, which became both an incredible honour and a fundamental phase in my personal and professional development. I would also like to thank my co-supervisor, Dr. Ana Paula Mucha, to whom I thank for the opportunity of integrating her laboratory, where I was always given all the conditions to develop my work to the fullest of its potential. Secondly, I would like to acknowledge CIIMAR - Interdisciplinary Centre of Marine and Environmental Research and Departamento de Química e Bioquímica of Faculty of Sciences of University of Porto, for the use of all the equipment, installations and facilities. To my lab mates at Ecobiotec (CIIMAR-UP), I recognise their friendship, as well as all the input in my work and precious help and support, with a special emphasis to Patricia Duarte, Filipa Santos, Joana Fernandes and Inês Ribeiro. -

University of Groningen Labrys Portucalensis Sp Nov., A
University of Groningen Labrys portucalensis sp nov., a fluorobenzene-degrading bacterium isolated from an industrially contaminated sediment in northern Portugal Carvalho, Maria F.; De Marco, Paolo; Duque, Anouk F.; Pacheco, Catarina C.; Janssen, Dick; Castro, Paula M. L. Published in: International Journal of Systematic and Evolutionary Microbiology DOI: 10.1099/ijs.0.65472-0 IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2008 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Carvalho, M. F., De Marco, P., Duque, A. F., Pacheco, C. C., Janssen, D. B., & Castro, P. M. L. (2008). Labrys portucalensis sp nov., a fluorobenzene-degrading bacterium isolated from an industrially contaminated sediment in northern Portugal. International Journal of Systematic and Evolutionary Microbiology, 58(3), 692-698. DOI: 10.1099/ijs.0.65472-0 Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons the number of authors shown on this cover page is limited to 10 maximum. -

Tree-Aggregated Predictive Modeling of Microbiome Data
www.nature.com/scientificreports OPEN Tree‑aggregated predictive modeling of microbiome data Jacob Bien1, Xiaohan Yan2, Léo Simpson3,4 & Christian L. Müller4,5,6* Modern high‑throughput sequencing technologies provide low‑cost microbiome survey data across all habitats of life at unprecedented scale. At the most granular level, the primary data consist of sparse counts of amplicon sequence variants or operational taxonomic units that are associated with taxonomic and phylogenetic group information. In this contribution, we leverage the hierarchical structure of amplicon data and propose a data‑driven and scalable tree‑guided aggregation framework to associate microbial subcompositions with response variables of interest. The excess number of zero or low count measurements at the read level forces traditional microbiome data analysis workfows to remove rare sequencing variants or group them by a fxed taxonomic rank, such as genus or phylum, or by phylogenetic similarity. By contrast, our framework, which we call trac (tree‑aggregation of compositional data), learns data‑adaptive taxon aggregation levels for predictive modeling, greatly reducing the need for user‑defned aggregation in preprocessing while simultaneously integrating seamlessly into the compositional data analysis framework. We illustrate the versatility of our framework in the context of large‑scale regression problems in human gut, soil, and marine microbial ecosystems. We posit that the inferred aggregation levels provide highly interpretable taxon groupings that can help microbiome researchers gain insights into the structure and functioning of the underlying ecosystem of interest. Microbial communities populate all major environments on earth and signifcantly contribute to the total plan- etary biomass. Current estimates suggest that a typical human-associated microbiome consists of ∼ 1013 bacteria1 and that marine bacteria and protists contribute to as much as 70% of the total marine biomass2. -

Pró-Reitoria De Pós-Graduação E Pesquisa Stricto Sensu Em Ciências Genômicas E Biotecnologia
Pró-Reitoria de Pós-Graduação e Pesquisa Stricto Sensu em Ciências Genômicas e Biotecnologia IDENTIFICAÇÃO, ISOLAMENTO E CARACTERIZAÇÃO DE BACTÉRIAS DE SOLO DE CERRADO PERTENCENTES AO FILO ACIDOBACTERIA Autor: Virgilio Hipólito Lemos de Castro Orientadora: Drª Cristine Chaves Barreto Brasília - DF 2011 VIRGILIO HIPÓLITO LEMOS DE CASTRO IDENTIFICAÇÃO, ISOLAMENTO E CARACTERIZAÇÃO DE BACTÉRIAS DE SOLO DE CERRADO PERTENCENTES AO FILO ACIDOBACTERIA Dissertação apresentada ao Programa de Pós- Graduação Stricto Sensu em Ciências Genômicas e Biotecnologia da Universidade Católica de Brasília, como requisito para obtenção do Título de Mestre em Ciências Genômicas e Biotecnologia. Orientadora: Drª. Cristine Chaves Barreto Brasília 2011 C355i Castro, Virgilio Hipólito Lemos de. Identificação, isolamento e caracterização de bactérias de solo de cerrado pertencentes ao filo acidobactéria – 2011. 76f. : il.; 30 cm Dissertação (mestrado – Universidade Católica de Brasília, 2011). Orientação: Cristine Chaves Barreto Ficha1. elaborada Bactéria. 2.pela Composição Biblioteca doPós solo.-Graduação 3. Biotecnologia. da UCB I. Barreto , Cristine Chaves, orient. II. Título. CDU 579 Dissertação de autoria de Virgilio Hipólito Lemos de Castro, intitulada “IDENTIFICAÇÃO, ISOLAMENTO E CARACTERIZAÇÃO DE BACTÉRIAS DE SOLO DE CERRADO PERTENCENTES AO FILO ACIDOBACTERIA”, apresentada como requisito para obtenção do grau de Mestre em Ciências Genômicas e Biotecnologia da Universidade Católica de Brasília em 27 de junho, defendida e aprovada pela banca examinadora abaixo -

Microbial Community Analysis in Soil (Rhizosphere) and the Marine (Plastisphere) Environment in Function of Plant Health and Biofilm Formation
Microbial community analysis in soil (rhizosphere) and the marine (plastisphere) environment in function of plant health and biofilm formation Caroline De Tender Thesis submitted in fulfillment of the requirements for the degree of Doctor (PhD) in Biotechnology Promotors: Prof. Dr. Peter Dawyndt Department of Applied mathematics, Computer Science and Statistics Faculty of Science Ghent University Dr. Martine Maes Crop Protection - Plant Sciences Unit Institute for Agricultural, Fisheries and Food Research (ILVO) Ir. Lisa Devriese Fisheries – Animal Sciences Unit Insitute for Agricultural, Fisheries and Food Research (ILVO) Dank je wel! De allerlaatste woorden die geschreven worden voor deze thesis zijn waarschijnlijk de eerste die gelezen worden door velen. Ongeveer vier jaar geleden startte ik mijn doctoraat bij het ILVO. Met volle moed begon ik aan mijn avontuur. Het ging niet altijd even vlot en ik kan eerlijk bekennen dat meerdere grenzen verlegd zijn. Vooral de combinatie van twee onderwerpen bleek niet altijd evident en kostte me meer dan eens bloed, zweet en tranen. Daarentegen bracht het ook vele opportuniteiten. De enige dag kon ik aan het wroeten zijn in de serre, terwijl ik de dag erop op de Simon Stevin sprong (en dit mag letterlijk worden genomen!) om plastic uit zee te vissen. Ja, het was me wel het avontuur… Natuurlijk zou dit allemaal niet mogelijk geweest zijn zonder de hulp van een aantal geweldige mensen. In de eerste plaats, mijn promotoren: Prof. Peter Dawyndt, dr. Martine Maes en natuurlijk Lisa Devriese. Dank je wel om vier jaar geleden het vertrouwen te hebben om mij dit onderzoek toe te wijzen, me steeds in de juiste richting te duwen als ik het Noorden even kwijt was, maar ook voor de gezellige babbels op de bureau. -

Microbial Communities of Polymetallic Deposits' Acidic
Microbial Communities of Polymetallic Deposits’ Acidic Ecosystems of ANGOR UNIVERSITY Continental Climatic Zone With High Temperature Contrasts Gavrilov, Sergei N.; Korzhenkov, Aleksei A.; Kublanov, Ilya V.; Bargiela, Rafael; Zamana, Leonid V.; Toshchakov, Stepan V.; Golyshin, Peter; Golyshina, Olga Frontiers in Microbiology PRIFYSGOL BANGOR / B Published: 17/07/2019 Publisher's PDF, also known as Version of record Cyswllt i'r cyhoeddiad / Link to publication Dyfyniad o'r fersiwn a gyhoeddwyd / Citation for published version (APA): Gavrilov, S. N., Korzhenkov, A. A., Kublanov, I. V., Bargiela, R., Zamana, L. V., Toshchakov, S. V., Golyshin, P., & Golyshina, O. (2019). Microbial Communities of Polymetallic Deposits’ Acidic Ecosystems of Continental Climatic Zone With High Temperature Contrasts. Frontiers in Microbiology. Hawliau Cyffredinol / General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. 01. Oct. 2021 ORIGINAL RESEARCH published: 17 July 2019 doi: 10.3389/fmicb.2019.01573 Microbial Communities of Polymetallic Deposits’ Acidic Ecosystems of Continental Climatic Zone With High Temperature Contrasts Sergey N. -
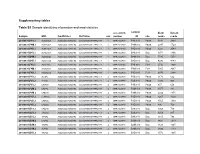
Supplementary Tables Table S1 Sample Identifying Information And
Supplementary tables Table S1 Sample identifying information and read statistics accession sample #raw #clean Sample MID FwdPrimer RvPrimer run number ID site reads reads 20100831.P4S.1 ACACGAGA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 P4S.910 Palsa 3831 2893 20100831.P4M.1 ACATAGCA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 P4M.910 Palsa 2047 1729 20100831.P4D.1 ACGATACA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 P4D.910 Palsa 3221 2543 20100831.B4S.1 ACTGCTCA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 B4S.910 Bog 1971 1366 20100831.B4M.1 AGCAGCGC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 B4M.910 Bog 5530 5251 20100831.B4D.1 AGCGTGCA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 B4D.910 Bog 4296 4143 20100831.F4S.1 AGTCTAGC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 F4S.910 Fen* 2716 1890 20100831.F4M.1 ATGACTGA ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 F4M.910 Fen* 5005 3987 20100831.F4D.1 ATGCACGC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 1 SRR1023739 F4D.910 Fen* 2870 2384 20100901.P1S.2 ACTGAT ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P1S.910 Palsa 1170 662 20100901.P2S.2 ATGTGT ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P2S.910 Palsa 1320 923 20100901.P3S.2 CACAGT ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P3S.910 Palsa 857 656 20100901.P2M.2 CACTAC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P2M.910 Palsa 1077 881 20100901.P3M.2 CAGCAT ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT 2 SRR1023740 P3M.910 Palsa 2032 1474 20100901.P1D.2 CTAGAGC ACGGGCGGTGWGTRC CCGYCAATTCMTTTRAGTTT